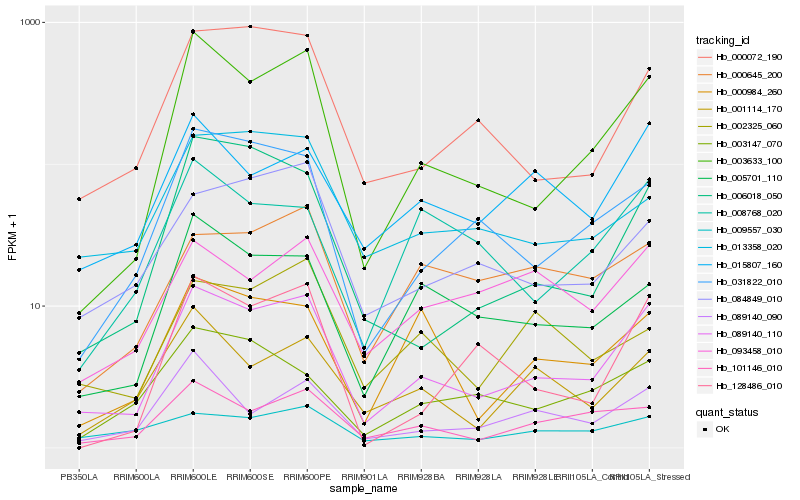
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_089140_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003633_100 |
0.1138905398 |
- |
- |
hypoia-responsive family protein 1 [Hevea brasiliensis] |
| 3 |
Hb_000072_190 |
0.1524061829 |
- |
- |
putative S-adenosylmethionine decarboxylase [Prunus dulcis] |
| 4 |
Hb_031822_010 |
0.1564528721 |
- |
- |
PREDICTED: uncharacterized protein LOC103496015 [Cucumis melo] |
| 5 |
Hb_101146_010 |
0.1626497261 |
- |
- |
PREDICTED: peregrin-like [Jatropha curcas] |
| 6 |
Hb_005701_110 |
0.1691987735 |
- |
- |
PREDICTED: probable 1-acylglycerol-3-phosphate O-acyltransferase [Jatropha curcas] |
| 7 |
Hb_006018_050 |
0.1703647866 |
- |
- |
PREDICTED: probable protein phosphatase 2C 63 [Jatropha curcas] |
| 8 |
Hb_084849_010 |
0.1704099654 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_015807_160 |
0.1735586091 |
- |
- |
PREDICTED: probable histone H2A.1 [Jatropha curcas] |
| 10 |
Hb_013358_020 |
0.1777448387 |
- |
- |
ccr4-associated factor, putative [Ricinus communis] |
| 11 |
Hb_089140_090 |
0.1781748472 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X3 [Beta vulgaris subsp. vulgaris] |
| 12 |
Hb_001114_170 |
0.1800661953 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_128486_010 |
0.1807647339 |
- |
- |
PREDICTED: U-box domain-containing protein 28 [Jatropha curcas] |
| 14 |
Hb_000984_260 |
0.1810564834 |
- |
- |
ergosterol biosynthetic protein 28 [Jatropha curcas] |
| 15 |
Hb_009557_030 |
0.1920119817 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-52-like [Jatropha curcas] |
| 16 |
Hb_003147_070 |
0.1927693609 |
transcription factor |
TF Family: NAC |
PREDICTED: transcription factor JUNGBRUNNEN 1-like [Jatropha curcas] |
| 17 |
Hb_093458_010 |
0.1939354098 |
- |
- |
hypothetical protein CISIN_1g0277592mg [Citrus sinensis] |
| 18 |
Hb_000645_200 |
0.1943408632 |
- |
- |
PREDICTED: GTP-binding protein SAR1A [Jatropha curcas] |
| 19 |
Hb_008768_020 |
0.1943944385 |
- |
- |
PREDICTED: uncharacterized protein LOC105639378 [Jatropha curcas] |
| 20 |
Hb_002325_060 |
0.1954093166 |
- |
- |
PREDICTED: ADP-ribosylation factor-like protein 8B-like [Glycine max] |