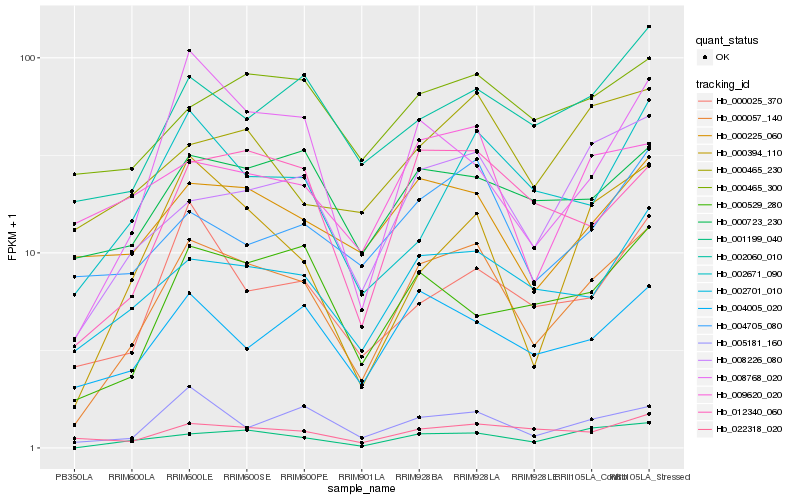
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000057_140 |
0.0 |
- |
- |
Mitochondrial carnitine/acylcarnitine carrier protein, putative [Ricinus communis] |
| 2 |
Hb_001199_040 |
0.1321722623 |
- |
- |
unnamed protein product [Coffea canephora] |
| 3 |
Hb_008226_080 |
0.1439293831 |
- |
- |
Protein LRP16, putative [Ricinus communis] |
| 4 |
Hb_012340_060 |
0.1456892132 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002701_010 |
0.1461842018 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 6 |
Hb_005181_160 |
0.1567678618 |
- |
- |
PREDICTED: uncharacterized protein LOC105643546 [Jatropha curcas] |
| 7 |
Hb_004005_020 |
0.1619647903 |
- |
- |
PREDICTED: nudix hydrolase 20, chloroplastic-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000394_110 |
0.1627923076 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 9 |
Hb_000225_060 |
0.1673804074 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase-like [Jatropha curcas] |
| 10 |
Hb_000025_370 |
0.168001839 |
- |
- |
PREDICTED: uncharacterized protein LOC105628992 isoform X1 [Jatropha curcas] |
| 11 |
Hb_008768_020 |
0.1687339159 |
- |
- |
PREDICTED: uncharacterized protein LOC105639378 [Jatropha curcas] |
| 12 |
Hb_022318_020 |
0.1691504731 |
- |
- |
hypothetical protein JCGZ_00541 [Jatropha curcas] |
| 13 |
Hb_002060_010 |
0.169494379 |
- |
- |
PREDICTED: uncharacterized protein LOC105642435 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000723_230 |
0.1701690121 |
- |
- |
hypothetical protein CISIN_1g017413mg [Citrus sinensis] |
| 15 |
Hb_000529_280 |
0.1704822326 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_002671_090 |
0.1727851151 |
- |
- |
PREDICTED: guanylate kinase 2 [Jatropha curcas] |
| 17 |
Hb_004705_080 |
0.1768334129 |
transcription factor |
TF Family: ARF |
transcription factor, putative [Ricinus communis] |
| 18 |
Hb_000465_230 |
0.178777764 |
- |
- |
PREDICTED: transcriptional regulator ATRX homolog [Jatropha curcas] |
| 19 |
Hb_000465_300 |
0.1788792202 |
- |
- |
PREDICTED: putative dual specificity protein phosphatase DSP8 [Jatropha curcas] |
| 20 |
Hb_009620_020 |
0.1794750973 |
- |
- |
conserved hypothetical protein [Ricinus communis] |