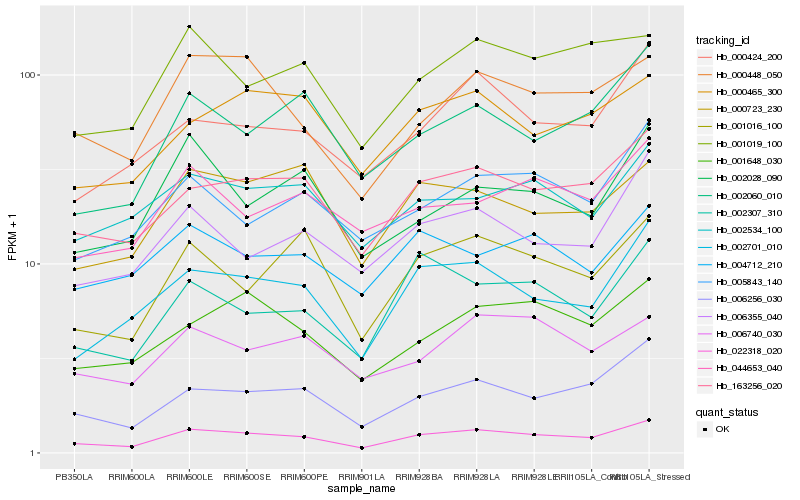
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_022318_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_00541 [Jatropha curcas] |
| 2 |
Hb_163256_020 |
0.0778658045 |
- |
- |
fructokinase [Manihot esculenta] |
| 3 |
Hb_002701_010 |
0.0874861461 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 4 |
Hb_006256_030 |
0.0902569185 |
- |
- |
PREDICTED: putative cyclic nucleotide-gated ion channel 18 [Jatropha curcas] |
| 5 |
Hb_000448_050 |
0.1005157683 |
- |
- |
PREDICTED: ubiquinol oxidase, mitochondrial [Jatropha curcas] |
| 6 |
Hb_001648_030 |
0.1028058336 |
- |
- |
PREDICTED: uncharacterized protein LOC105628931 isoform X2 [Jatropha curcas] |
| 7 |
Hb_001016_100 |
0.1061257064 |
- |
- |
PREDICTED: V-type proton ATPase subunit E-like [Pyrus x bretschneideri] |
| 8 |
Hb_002307_310 |
0.1088851843 |
- |
- |
PREDICTED: outer envelope pore protein 37, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000723_230 |
0.1094569005 |
- |
- |
hypothetical protein CISIN_1g017413mg [Citrus sinensis] |
| 10 |
Hb_001019_100 |
0.1106750097 |
- |
- |
BnaC02g13240D [Brassica napus] |
| 11 |
Hb_000465_300 |
0.1112603498 |
- |
- |
PREDICTED: putative dual specificity protein phosphatase DSP8 [Jatropha curcas] |
| 12 |
Hb_000424_200 |
0.1114951794 |
- |
- |
hypothetical protein JCGZ_07583 [Jatropha curcas] |
| 13 |
Hb_002534_100 |
0.1118191156 |
- |
- |
PREDICTED: malate dehydrogenase, chloroplastic [Jatropha curcas] |
| 14 |
Hb_006355_040 |
0.1138683697 |
- |
- |
PREDICTED: chaperonin CPN60-2, mitochondrial-like [Jatropha curcas] |
| 15 |
Hb_006740_030 |
0.115731035 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 16 |
Hb_005843_140 |
0.1162377376 |
- |
- |
Red chlorophyll catabolite reductase, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_044653_040 |
0.1175443127 |
transcription factor |
TF Family: GNAT |
PREDICTED: N-alpha-acetyltransferase 11 [Jatropha curcas] |
| 18 |
Hb_002060_010 |
0.1179118336 |
- |
- |
PREDICTED: uncharacterized protein LOC105642435 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004712_210 |
0.1188888124 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM50-like [Jatropha curcas] |
| 20 |
Hb_002028_090 |
0.1227399507 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase AIP2-like [Jatropha curcas] |