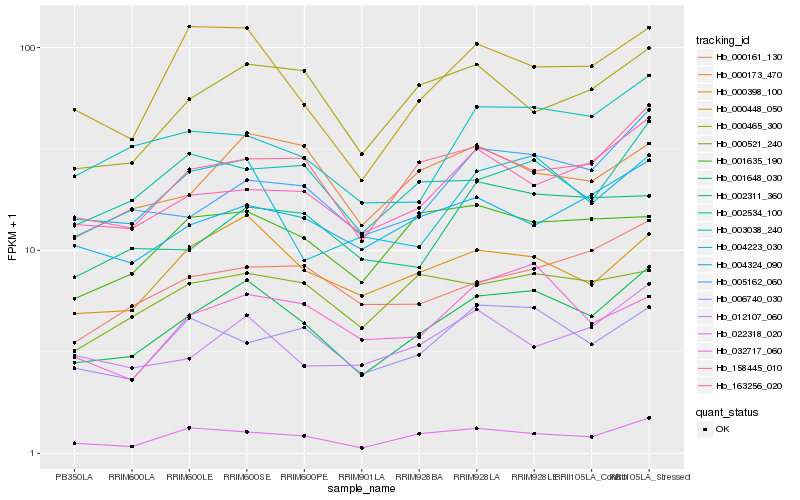
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001648_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628931 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000398_100 |
0.0934248356 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 3 |
Hb_163256_020 |
0.0940064541 |
- |
- |
fructokinase [Manihot esculenta] |
| 4 |
Hb_000448_050 |
0.1021003334 |
- |
- |
PREDICTED: ubiquinol oxidase, mitochondrial [Jatropha curcas] |
| 5 |
Hb_005162_060 |
0.1026720321 |
- |
- |
PREDICTED: putative septum site-determining protein minD homolog, chloroplastic [Jatropha curcas] |
| 6 |
Hb_022318_020 |
0.1028058336 |
- |
- |
hypothetical protein JCGZ_00541 [Jatropha curcas] |
| 7 |
Hb_002534_100 |
0.1080566139 |
- |
- |
PREDICTED: malate dehydrogenase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000173_470 |
0.1090542004 |
- |
- |
PREDICTED: BSD domain-containing protein 1 [Jatropha curcas] |
| 9 |
Hb_000465_300 |
0.10969666 |
- |
- |
PREDICTED: putative dual specificity protein phosphatase DSP8 [Jatropha curcas] |
| 10 |
Hb_003038_240 |
0.1110582233 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_158445_010 |
0.1129458529 |
- |
- |
PREDICTED: cysteine protease ATG4-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_006740_030 |
0.1142253349 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 13 |
Hb_004324_090 |
0.1142646972 |
- |
- |
PREDICTED: uncharacterized protein LOC105648352 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000161_130 |
0.1158105662 |
- |
- |
PREDICTED: nifU-like protein 2, chloroplastic [Jatropha curcas] |
| 15 |
Hb_004223_030 |
0.1167316669 |
- |
- |
PREDICTED: uncharacterized protein LOC105635347 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000521_240 |
0.1171796716 |
- |
- |
PREDICTED: uncharacterized protein LOC105650339 isoform X2 [Jatropha curcas] |
| 17 |
Hb_012107_060 |
0.1178849314 |
- |
- |
hypothetical protein JCGZ_17178 [Jatropha curcas] |
| 18 |
Hb_001635_190 |
0.1179359898 |
- |
- |
RNA m5u methyltransferase, putative [Ricinus communis] |
| 19 |
Hb_002311_360 |
0.1193601969 |
- |
- |
PREDICTED: uncharacterized protein LOC105114273 [Populus euphratica] |
| 20 |
Hb_032717_060 |
0.1203951642 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ18 isoform X1 [Jatropha curcas] |