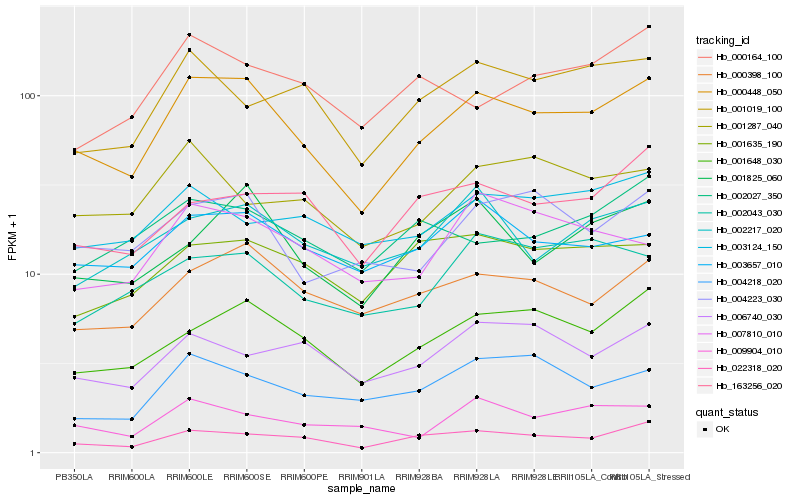
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000448_050 |
0.0 |
- |
- |
PREDICTED: ubiquinol oxidase, mitochondrial [Jatropha curcas] |
| 2 |
Hb_022318_020 |
0.1005157683 |
- |
- |
hypothetical protein JCGZ_00541 [Jatropha curcas] |
| 3 |
Hb_001648_030 |
0.1021003334 |
- |
- |
PREDICTED: uncharacterized protein LOC105628931 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000398_100 |
0.1112298954 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 5 |
Hb_004223_030 |
0.1176549334 |
- |
- |
PREDICTED: uncharacterized protein LOC105635347 isoform X1 [Jatropha curcas] |
| 6 |
Hb_003657_010 |
0.1208088045 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001825_060 |
0.1217410057 |
- |
- |
PREDICTED: F-box/kelch-repeat protein OR23 [Jatropha curcas] |
| 8 |
Hb_001019_100 |
0.1277203903 |
- |
- |
BnaC02g13240D [Brassica napus] |
| 9 |
Hb_002217_020 |
0.1283405186 |
- |
- |
PREDICTED: uncharacterized protein LOC105643601 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001635_190 |
0.1309038288 |
- |
- |
RNA m5u methyltransferase, putative [Ricinus communis] |
| 11 |
Hb_007810_010 |
0.1317233121 |
- |
- |
PREDICTED: uncharacterized protein LOC105647655 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002043_030 |
0.1320929372 |
- |
- |
hypothetical protein RCOM_1486170 [Ricinus communis] |
| 13 |
Hb_003124_150 |
0.1325134398 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 28-like [Jatropha curcas] |
| 14 |
Hb_009904_010 |
0.1325880294 |
- |
- |
hypothetical protein JCGZ_16885 [Jatropha curcas] |
| 15 |
Hb_004218_020 |
0.1335107638 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_163256_020 |
0.1336275085 |
- |
- |
fructokinase [Manihot esculenta] |
| 17 |
Hb_001287_040 |
0.1341859117 |
- |
- |
PREDICTED: kinesin-like protein NACK1 [Jatropha curcas] |
| 18 |
Hb_002027_350 |
0.1354803235 |
- |
- |
PREDICTED: U1 small nuclear ribonucleoprotein C-like [Jatropha curcas] |
| 19 |
Hb_006740_030 |
0.136882658 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 20 |
Hb_000164_100 |
0.1389090724 |
- |
- |
PREDICTED: probable histone H2A.1 [Jatropha curcas] |