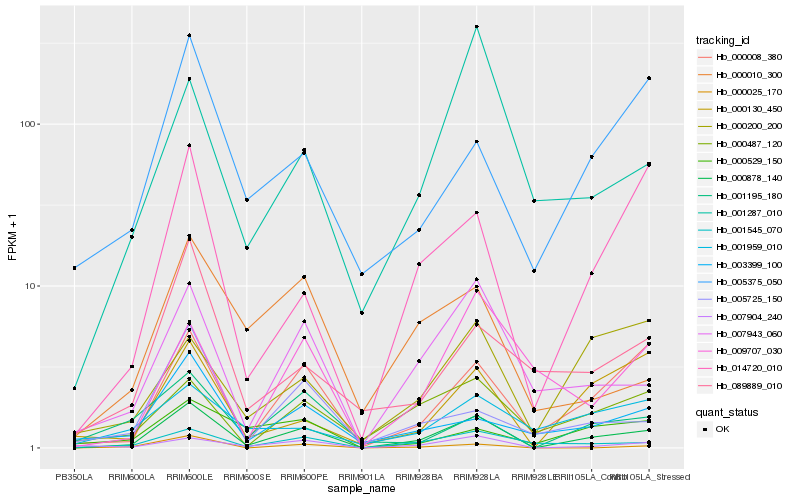
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000878_140 |
0.0 |
- |
- |
PREDICTED: cell division cycle 20.2, cofactor of APC complex-like [Jatropha curcas] |
| 2 |
Hb_000008_380 |
0.1946005205 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_001545_070 |
0.2161839112 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_007943_060 |
0.2179817995 |
- |
- |
Rop5 [Hevea brasiliensis] |
| 5 |
Hb_007904_240 |
0.2237615741 |
- |
- |
PREDICTED: protein terminal ear1 [Jatropha curcas] |
| 6 |
Hb_014720_010 |
0.244608489 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000487_120 |
0.2469266498 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000130_450 |
0.250745484 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000025_170 |
0.2531233597 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003399_100 |
0.2569848718 |
- |
- |
PREDICTED: putative cyclin-A3-1 [Jatropha curcas] |
| 11 |
Hb_001195_180 |
0.2613190245 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 5 [Jatropha curcas] |
| 12 |
Hb_005375_050 |
0.2650284781 |
- |
- |
inorganic pyrophosphatase, putative [Ricinus communis] |
| 13 |
Hb_089889_010 |
0.2685887973 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 [Jatropha curcas] |
| 14 |
Hb_005725_150 |
0.2711439653 |
- |
- |
PREDICTED: kinesin-related protein 11 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000200_200 |
0.2752898872 |
- |
- |
PREDICTED: putative zinc finger protein At1g68190 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000010_300 |
0.2758143577 |
transcription factor |
TF Family: TAZ |
protein binding protein, putative [Ricinus communis] |
| 17 |
Hb_001287_010 |
0.278868283 |
- |
- |
PREDICTED: arginine decarboxylase [Jatropha curcas] |
| 18 |
Hb_009707_030 |
0.2798280968 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620 [Jatropha curcas] |
| 19 |
Hb_001959_010 |
0.2798755764 |
- |
- |
myosin XI, putative [Ricinus communis] |
| 20 |
Hb_000529_150 |
0.2817956196 |
- |
- |
protein with unknown function [Ricinus communis] |