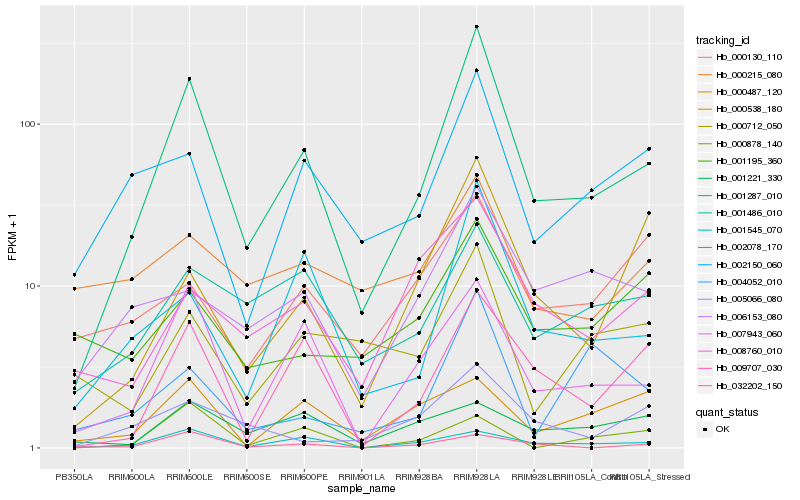
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001287_010 |
0.0 |
- |
- |
PREDICTED: arginine decarboxylase [Jatropha curcas] |
| 2 |
Hb_007943_060 |
0.1856947794 |
- |
- |
Rop5 [Hevea brasiliensis] |
| 3 |
Hb_009707_030 |
0.2052929565 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620 [Jatropha curcas] |
| 4 |
Hb_002078_170 |
0.208111684 |
- |
- |
ELMO/CED-12 domain-containing protein isoform 3 [Theobroma cacao] |
| 5 |
Hb_001545_070 |
0.2183432733 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002150_060 |
0.2318112881 |
- |
- |
PREDICTED: arabinogalactan peptide 20-like [Jatropha curcas] |
| 7 |
Hb_000538_180 |
0.2439390718 |
- |
- |
PREDICTED: probable polyamine oxidase 4 [Jatropha curcas] |
| 8 |
Hb_001486_010 |
0.2479926877 |
- |
- |
adenylsulfate kinase, putative [Ricinus communis] |
| 9 |
Hb_006153_080 |
0.251441304 |
- |
- |
flavin-containing monooxygenase-related family protein [Populus trichocarpa] |
| 10 |
Hb_008760_010 |
0.2530623763 |
- |
- |
berberine bridge enzyme [Hevea brasiliensis] |
| 11 |
Hb_032202_150 |
0.254303294 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR14 [Jatropha curcas] |
| 12 |
Hb_005066_080 |
0.2544210072 |
- |
- |
hypothetical protein PRUPE_ppa025081mg [Prunus persica] |
| 13 |
Hb_000712_050 |
0.263738768 |
- |
- |
PREDICTED: inositol oxygenase 1-like isoform X1 [Citrus sinensis] |
| 14 |
Hb_004052_010 |
0.268321918 |
- |
- |
PREDICTED: ninja-family protein AFP3-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_001195_360 |
0.2776220228 |
- |
- |
PREDICTED: random slug protein 5 [Jatropha curcas] |
| 16 |
Hb_000878_140 |
0.278868283 |
- |
- |
PREDICTED: cell division cycle 20.2, cofactor of APC complex-like [Jatropha curcas] |
| 17 |
Hb_000487_120 |
0.2789296458 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000215_080 |
0.2829949455 |
- |
- |
PREDICTED: uncharacterized protein LOC105637840 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000130_110 |
0.2834395079 |
- |
- |
PREDICTED: TPR repeat-containing thioredoxin TTL1 [Jatropha curcas] |
| 20 |
Hb_001221_330 |
0.28702889 |
- |
- |
PREDICTED: uncharacterized protein LOC105648791 [Jatropha curcas] |