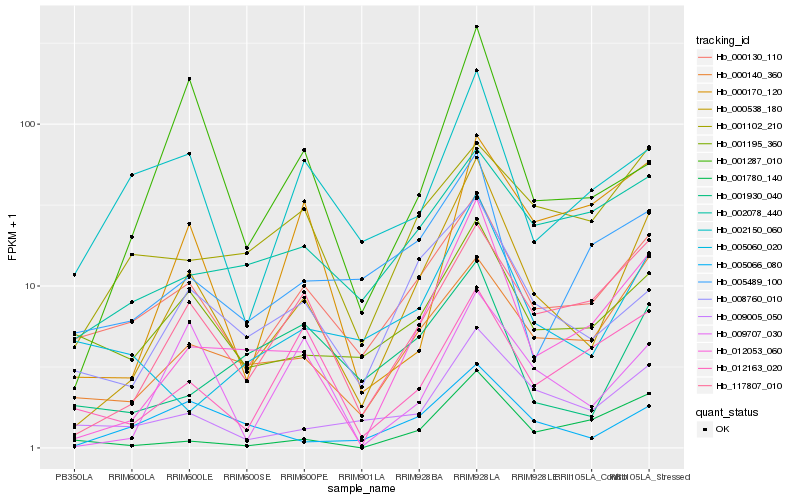
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000538_180 |
0.0 |
- |
- |
PREDICTED: probable polyamine oxidase 4 [Jatropha curcas] |
| 2 |
Hb_012053_060 |
0.1532859183 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g44820 isoform X2 [Jatropha curcas] |
| 3 |
Hb_117807_010 |
0.1883402037 |
- |
- |
PREDICTED: uncharacterized protein LOC105635720 [Jatropha curcas] |
| 4 |
Hb_000130_110 |
0.2004135946 |
- |
- |
PREDICTED: TPR repeat-containing thioredoxin TTL1 [Jatropha curcas] |
| 5 |
Hb_000140_360 |
0.2085393136 |
- |
- |
PREDICTED: uncharacterized protein LOC105642906 [Jatropha curcas] |
| 6 |
Hb_008760_010 |
0.2088713335 |
- |
- |
berberine bridge enzyme [Hevea brasiliensis] |
| 7 |
Hb_009005_050 |
0.2146432596 |
- |
- |
neutral/alkaline invertase 3 [Hevea brasiliensis] |
| 8 |
Hb_005066_080 |
0.2294876648 |
- |
- |
hypothetical protein PRUPE_ppa025081mg [Prunus persica] |
| 9 |
Hb_001780_140 |
0.2307491303 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 10 |
Hb_001195_360 |
0.2324545066 |
- |
- |
PREDICTED: random slug protein 5 [Jatropha curcas] |
| 11 |
Hb_009707_030 |
0.2422100533 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620 [Jatropha curcas] |
| 12 |
Hb_001287_010 |
0.2439390718 |
- |
- |
PREDICTED: arginine decarboxylase [Jatropha curcas] |
| 13 |
Hb_005060_020 |
0.2450536316 |
- |
- |
PREDICTED: metal tolerance protein 10-like isoform X1 [Populus euphratica] |
| 14 |
Hb_005489_100 |
0.2533247913 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002150_060 |
0.2544178231 |
- |
- |
PREDICTED: arabinogalactan peptide 20-like [Jatropha curcas] |
| 16 |
Hb_001102_210 |
0.2552281329 |
- |
- |
PREDICTED: alpha,alpha-trehalose-phosphate synthase [UDP-forming] 1-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_002078_440 |
0.2563731896 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 3 isoform X1 [Jatropha curcas] |
| 18 |
Hb_012163_020 |
0.2578595175 |
- |
- |
PREDICTED: uncharacterized protein LOC105650815 isoform X3 [Jatropha curcas] |
| 19 |
Hb_001930_040 |
0.2595697766 |
- |
- |
PREDICTED: serine acetyltransferase 1, chloroplastic-like [Beta vulgaris subsp. vulgaris] |
| 20 |
Hb_000170_120 |
0.2599616161 |
- |
- |
ATP binding protein, putative [Ricinus communis] |