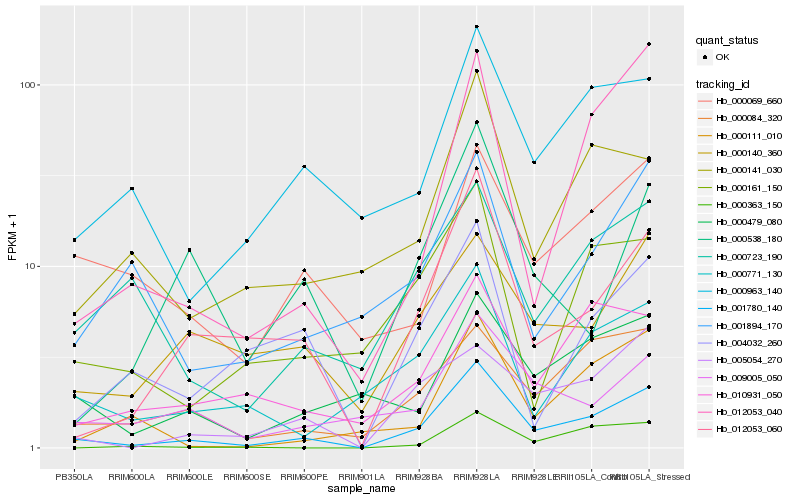
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001780_140 |
0.0 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 2 |
Hb_012053_060 |
0.1722949798 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g44820 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000084_320 |
0.2007852015 |
- |
- |
PREDICTED: probable small nuclear ribonucleoprotein G [Populus euphratica] |
| 4 |
Hb_005054_270 |
0.2028197156 |
- |
- |
- |
| 5 |
Hb_000771_130 |
0.2101115032 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 6 |
Hb_012053_040 |
0.2129589509 |
- |
- |
PREDICTED: putative phospholipid:diacylglycerol acyltransferase 2 [Jatropha curcas] |
| 7 |
Hb_000479_080 |
0.2135755432 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 8 |
Hb_000963_140 |
0.218161613 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 9 |
Hb_010931_050 |
0.2183384879 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25-like [Jatropha curcas] |
| 10 |
Hb_009005_050 |
0.2191717041 |
- |
- |
neutral/alkaline invertase 3 [Hevea brasiliensis] |
| 11 |
Hb_000140_360 |
0.219403083 |
- |
- |
PREDICTED: uncharacterized protein LOC105642906 [Jatropha curcas] |
| 12 |
Hb_000141_030 |
0.2206994202 |
- |
- |
PREDICTED: scarecrow-like protein 22 [Jatropha curcas] |
| 13 |
Hb_000111_010 |
0.2231796066 |
- |
- |
DNA-directed RNA polymerase, putative [Ricinus communis] |
| 14 |
Hb_000723_190 |
0.2236011169 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_000069_660 |
0.2247690443 |
- |
- |
PREDICTED: mitochondrial carrier protein MTM1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000363_150 |
0.2282383679 |
- |
- |
hypothetical protein POPTR_0011s09040g [Populus trichocarpa] |
| 17 |
Hb_000161_150 |
0.2282436721 |
- |
- |
PREDICTED: protein argonaute 2-like [Jatropha curcas] |
| 18 |
Hb_000538_180 |
0.2307491303 |
- |
- |
PREDICTED: probable polyamine oxidase 4 [Jatropha curcas] |
| 19 |
Hb_004032_260 |
0.2315513076 |
- |
- |
PREDICTED: uncharacterized protein LOC105635147 [Jatropha curcas] |
| 20 |
Hb_001894_170 |
0.2345087744 |
- |
- |
PREDICTED: pantothenate kinase 1 [Jatropha curcas] |