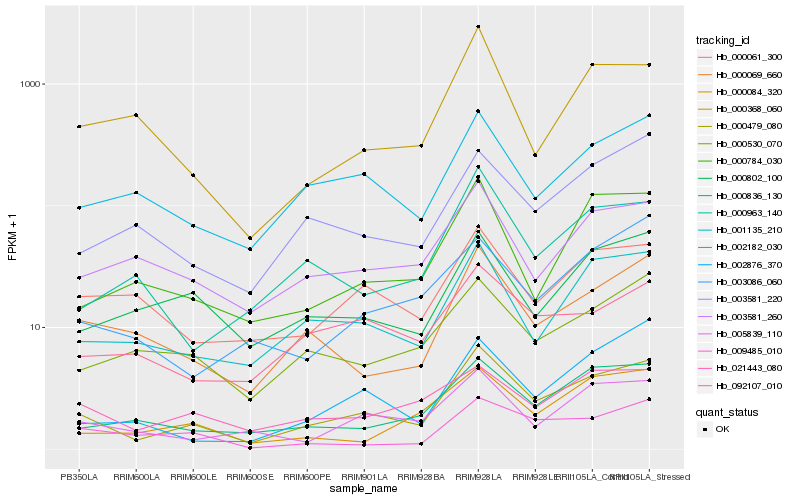
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000479_080 |
0.0 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 2 |
Hb_001135_210 |
0.1482043632 |
- |
- |
PREDICTED: probable adenylate kinase 6, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000836_130 |
0.1520833949 |
- |
- |
PREDICTED: uncharacterized protein LOC105642953 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000069_660 |
0.1582114102 |
- |
- |
PREDICTED: mitochondrial carrier protein MTM1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_005839_110 |
0.1634602864 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At5g47530-like [Populus euphratica] |
| 6 |
Hb_000784_030 |
0.1636304173 |
- |
- |
neutral/alkaline invertase 2 [Hevea brasiliensis] |
| 7 |
Hb_002182_030 |
0.1649579373 |
- |
- |
hypothetical protein Csa_5G139175 [Cucumis sativus] |
| 8 |
Hb_000530_070 |
0.1713456425 |
- |
- |
neutral/alkaline invertase 3 [Hevea brasiliensis] |
| 9 |
Hb_003581_220 |
0.1730938055 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_000963_140 |
0.1741442547 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 11 |
Hb_000084_320 |
0.174762292 |
- |
- |
PREDICTED: probable small nuclear ribonucleoprotein G [Populus euphratica] |
| 12 |
Hb_021443_080 |
0.174931049 |
- |
- |
PREDICTED: CDGSH iron-sulfur domain-containing protein NEET [Jatropha curcas] |
| 13 |
Hb_002876_370 |
0.1764815383 |
- |
- |
hypothetical protein B456_013G140400 [Gossypium raimondii] |
| 14 |
Hb_003581_260 |
0.1772010124 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 29-like [Jatropha curcas] |
| 15 |
Hb_000802_100 |
0.1774045209 |
- |
- |
PREDICTED: uncharacterized protein LOC105634009 [Jatropha curcas] |
| 16 |
Hb_092107_010 |
0.1796450308 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000368_060 |
0.1797723534 |
- |
- |
thioredoxin H-type 6 [Hevea brasiliensis] |
| 18 |
Hb_000061_300 |
0.1799295461 |
- |
- |
hypothetical protein JCGZ_11668 [Jatropha curcas] |
| 19 |
Hb_003086_060 |
0.1809666112 |
- |
- |
PREDICTED: uncharacterized protein LOC105629937 [Jatropha curcas] |
| 20 |
Hb_009485_010 |
0.1822314658 |
- |
- |
PREDICTED: uncharacterized protein LOC105641071 [Jatropha curcas] |