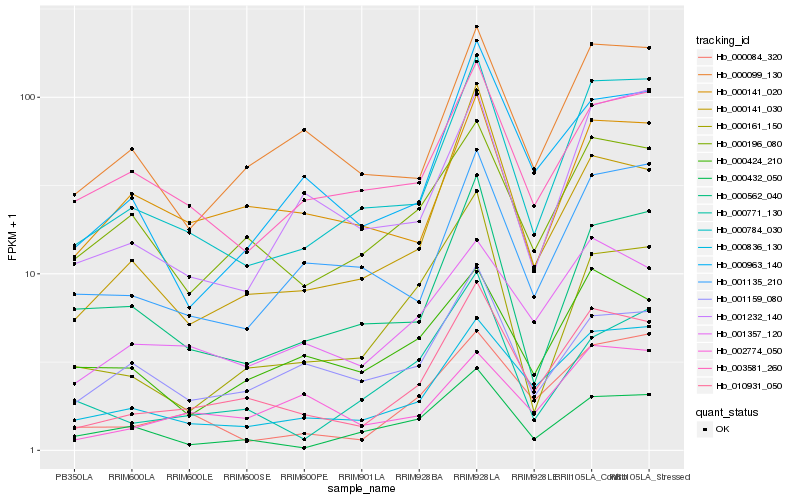
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010931_050 |
0.0 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25-like [Jatropha curcas] |
| 2 |
Hb_000836_130 |
0.120889912 |
- |
- |
PREDICTED: uncharacterized protein LOC105642953 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000784_030 |
0.1214326694 |
- |
- |
neutral/alkaline invertase 2 [Hevea brasiliensis] |
| 4 |
Hb_000141_030 |
0.1312901477 |
- |
- |
PREDICTED: scarecrow-like protein 22 [Jatropha curcas] |
| 5 |
Hb_001357_120 |
0.141490204 |
- |
- |
ubiquitin-activating enzyme E1, putative [Ricinus communis] |
| 6 |
Hb_000084_320 |
0.1423433465 |
- |
- |
PREDICTED: probable small nuclear ribonucleoprotein G [Populus euphratica] |
| 7 |
Hb_000963_140 |
0.1456871118 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 8 |
Hb_000099_130 |
0.1528548373 |
- |
- |
PREDICTED: uncharacterized protein LOC105645728 [Jatropha curcas] |
| 9 |
Hb_000196_080 |
0.155602049 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH79-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_001135_210 |
0.1688511003 |
- |
- |
PREDICTED: probable adenylate kinase 6, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001159_080 |
0.1694903455 |
- |
- |
hypothetical protein JCGZ_20745 [Jatropha curcas] |
| 12 |
Hb_002774_050 |
0.1710585659 |
- |
- |
PREDICTED: uncharacterized protein LOC105630601 [Jatropha curcas] |
| 13 |
Hb_001232_140 |
0.1740436584 |
- |
- |
PREDICTED: uncharacterized protein LOC105634966 [Jatropha curcas] |
| 14 |
Hb_000161_150 |
0.1748640159 |
- |
- |
PREDICTED: protein argonaute 2-like [Jatropha curcas] |
| 15 |
Hb_000562_040 |
0.1756635721 |
- |
- |
PREDICTED: auxin response factor 5 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000424_210 |
0.1759127702 |
- |
- |
PREDICTED: uncharacterized protein LOC105638063 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000141_020 |
0.1760993328 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 6 [Pyrus x bretschneideri] |
| 18 |
Hb_003581_260 |
0.1761637685 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 29-like [Jatropha curcas] |
| 19 |
Hb_000771_130 |
0.1763321638 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 20 |
Hb_000432_050 |
0.1765173615 |
- |
- |
GTP-binding protein typa/bipa, putative [Ricinus communis] |