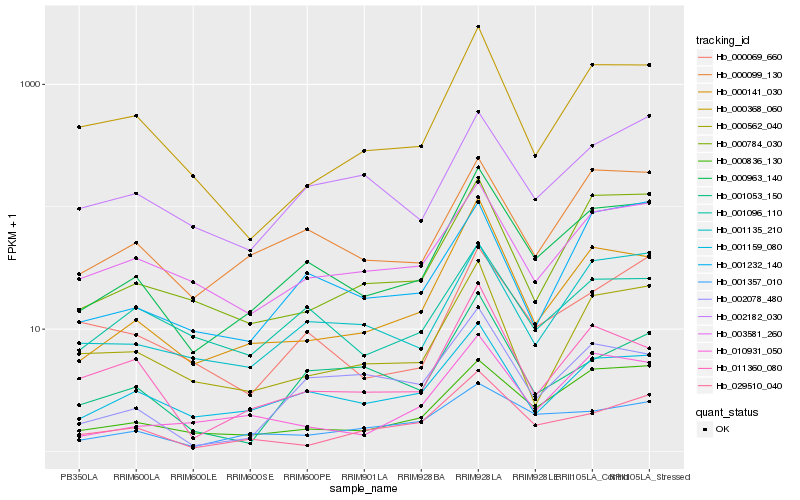
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000963_140 |
0.0 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 2 |
Hb_000099_130 |
0.1162241824 |
- |
- |
PREDICTED: uncharacterized protein LOC105645728 [Jatropha curcas] |
| 3 |
Hb_000141_030 |
0.1163197814 |
- |
- |
PREDICTED: scarecrow-like protein 22 [Jatropha curcas] |
| 4 |
Hb_001159_080 |
0.1217173068 |
- |
- |
hypothetical protein JCGZ_20745 [Jatropha curcas] |
| 5 |
Hb_000836_130 |
0.1261721289 |
- |
- |
PREDICTED: uncharacterized protein LOC105642953 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001135_210 |
0.1308467511 |
- |
- |
PREDICTED: probable adenylate kinase 6, chloroplastic [Jatropha curcas] |
| 7 |
Hb_003581_260 |
0.1429469189 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 29-like [Jatropha curcas] |
| 8 |
Hb_000784_030 |
0.1454174898 |
- |
- |
neutral/alkaline invertase 2 [Hevea brasiliensis] |
| 9 |
Hb_010931_050 |
0.1456871118 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25-like [Jatropha curcas] |
| 10 |
Hb_001096_110 |
0.1463757683 |
- |
- |
PREDICTED: probable VAMP-like protein At1g33475 [Jatropha curcas] |
| 11 |
Hb_002078_480 |
0.1466428111 |
- |
- |
hypothetical protein CICLE_v10033318mg [Citrus clementina] |
| 12 |
Hb_001232_140 |
0.1523705582 |
- |
- |
PREDICTED: uncharacterized protein LOC105634966 [Jatropha curcas] |
| 13 |
Hb_001357_010 |
0.152573527 |
- |
- |
- |
| 14 |
Hb_000069_660 |
0.1542346743 |
- |
- |
PREDICTED: mitochondrial carrier protein MTM1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_011360_080 |
0.1542927161 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_029510_040 |
0.1562714014 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g23330 [Vitis vinifera] |
| 17 |
Hb_000368_060 |
0.1565158841 |
- |
- |
thioredoxin H-type 6 [Hevea brasiliensis] |
| 18 |
Hb_001053_150 |
0.1566870685 |
- |
- |
- |
| 19 |
Hb_002182_030 |
0.1579186723 |
- |
- |
hypothetical protein Csa_5G139175 [Cucumis sativus] |
| 20 |
Hb_000562_040 |
0.1579295581 |
- |
- |
PREDICTED: auxin response factor 5 isoform X1 [Jatropha curcas] |