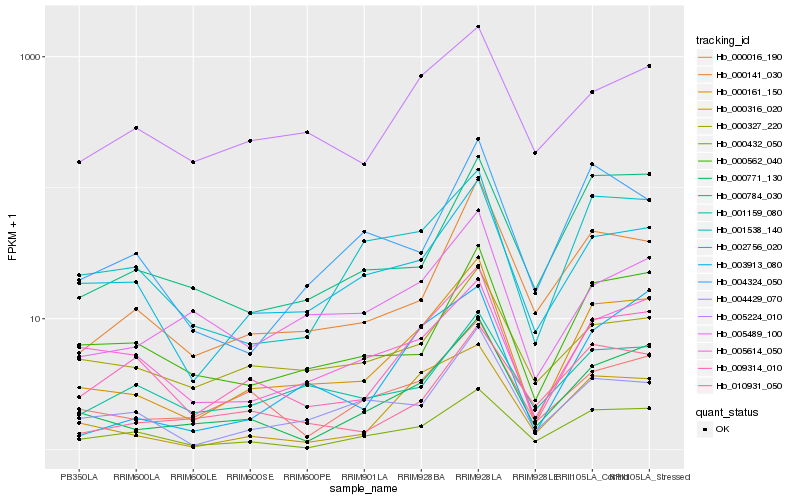
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000161_150 |
0.0 |
- |
- |
PREDICTED: protein argonaute 2-like [Jatropha curcas] |
| 2 |
Hb_009314_010 |
0.1096405938 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 3 |
Hb_000771_130 |
0.1158855373 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 4 |
Hb_002756_020 |
0.1376674556 |
transcription factor |
TF Family: HB |
bel1 homeotic protein, putative [Ricinus communis] |
| 5 |
Hb_000316_020 |
0.1398725217 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 6 |
Hb_000141_030 |
0.1467314563 |
- |
- |
PREDICTED: scarecrow-like protein 22 [Jatropha curcas] |
| 7 |
Hb_000432_050 |
0.1478305075 |
- |
- |
GTP-binding protein typa/bipa, putative [Ricinus communis] |
| 8 |
Hb_000562_040 |
0.1546768246 |
- |
- |
PREDICTED: auxin response factor 5 isoform X1 [Jatropha curcas] |
| 9 |
Hb_004429_070 |
0.1550438765 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 10 |
Hb_001538_140 |
0.1575701921 |
- |
- |
PREDICTED: PXMP2/4 family protein 4-like [Jatropha curcas] |
| 11 |
Hb_003913_080 |
0.1614034612 |
- |
- |
rpl27A [Hemiselmis andersenii] |
| 12 |
Hb_000016_190 |
0.1618978781 |
- |
- |
ABC transporter B family member 28 [Glycine soja] |
| 13 |
Hb_005489_100 |
0.1675896126 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_005224_010 |
0.1678715343 |
- |
- |
glycine-rich protein [Bruguiera gymnorhiza] |
| 15 |
Hb_000784_030 |
0.1705535657 |
- |
- |
neutral/alkaline invertase 2 [Hevea brasiliensis] |
| 16 |
Hb_005614_050 |
0.1707639765 |
- |
- |
PREDICTED: aspartate aminotransferase, mitochondrial-like [Jatropha curcas] |
| 17 |
Hb_000327_220 |
0.1718010597 |
- |
- |
PREDICTED: probable protein phosphatase 2C 38 [Jatropha curcas] |
| 18 |
Hb_010931_050 |
0.1748640159 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25-like [Jatropha curcas] |
| 19 |
Hb_001159_080 |
0.1754391937 |
- |
- |
hypothetical protein JCGZ_20745 [Jatropha curcas] |
| 20 |
Hb_004324_050 |
0.1807640781 |
- |
- |
PREDICTED: spermine synthase [Jatropha curcas] |