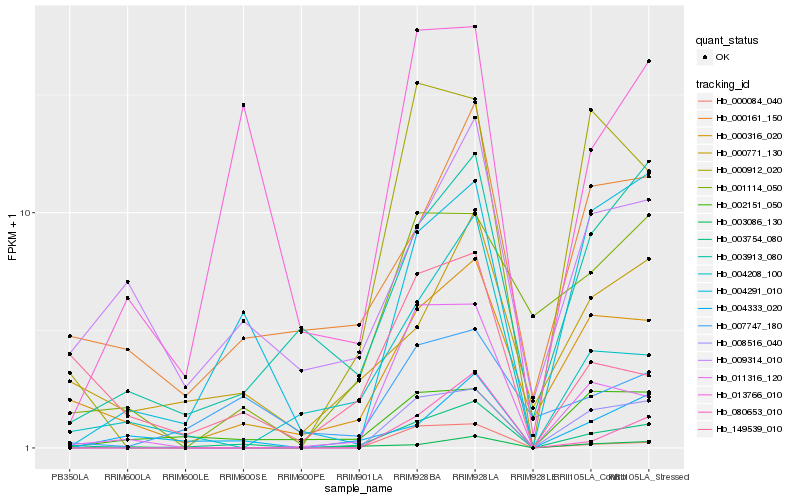
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003754_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 2 |
Hb_000316_020 |
0.1933714782 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 3 |
Hb_000161_150 |
0.2232215264 |
- |
- |
PREDICTED: protein argonaute 2-like [Jatropha curcas] |
| 4 |
Hb_000771_130 |
0.2232446874 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 5 |
Hb_008516_040 |
0.2267018681 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003913_080 |
0.233089201 |
- |
- |
rpl27A [Hemiselmis andersenii] |
| 7 |
Hb_004208_100 |
0.2355637799 |
- |
- |
Ribosomal protein L3 plastid isoform 1 [Theobroma cacao] |
| 8 |
Hb_004333_020 |
0.2419481346 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |
| 9 |
Hb_009314_010 |
0.2433395313 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 10 |
Hb_004291_010 |
0.2441320906 |
- |
- |
(R)-limonene synthase, putative [Ricinus communis] |
| 11 |
Hb_080653_010 |
0.2463959003 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_000912_020 |
0.2468978284 |
- |
- |
PREDICTED: uncharacterized protein LOC105645039 [Jatropha curcas] |
| 13 |
Hb_013766_010 |
0.2471822336 |
- |
- |
hypothetical protein JCGZ_00483 [Jatropha curcas] |
| 14 |
Hb_011316_120 |
0.2515280074 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH118-like [Jatropha curcas] |
| 15 |
Hb_007747_180 |
0.2545435245 |
transcription factor |
TF Family: TCP |
PREDICTED: bifunctional nitrilase/nitrile hydratase NIT4B-like [Jatropha curcas] |
| 16 |
Hb_149539_010 |
0.2556554025 |
- |
- |
- |
| 17 |
Hb_003086_130 |
0.2604146421 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 18 |
Hb_001114_050 |
0.2646101392 |
- |
- |
hypothetical protein RCOM_0744580 [Ricinus communis] |
| 19 |
Hb_002151_050 |
0.2646326595 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 20 |
Hb_000084_040 |
0.271257742 |
- |
- |
hypothetical protein RCOM_1007940 [Ricinus communis] |