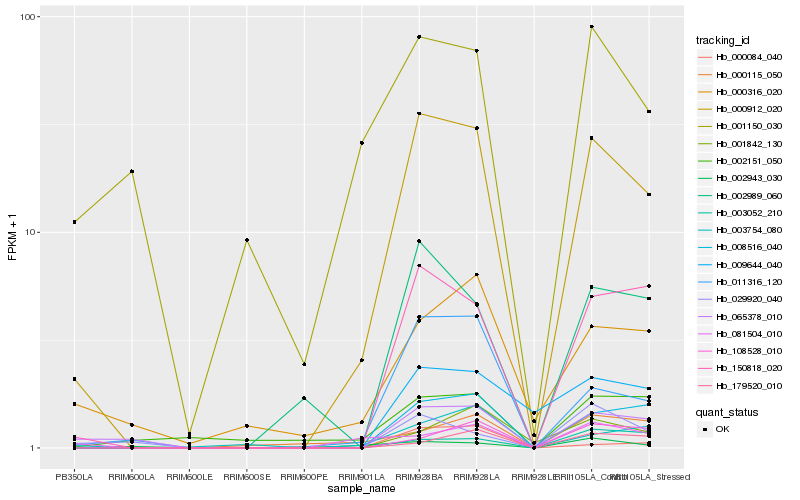
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000912_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645039 [Jatropha curcas] |
| 2 |
Hb_150818_020 |
0.1641533516 |
- |
- |
PREDICTED: uncharacterized protein LOC105645039 [Jatropha curcas] |
| 3 |
Hb_008516_040 |
0.1761387322 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_011316_120 |
0.1982322027 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH118-like [Jatropha curcas] |
| 5 |
Hb_002989_060 |
0.208685848 |
- |
- |
hypothetical protein JCGZ_22461 [Jatropha curcas] |
| 6 |
Hb_065378_010 |
0.2099600618 |
- |
- |
S-locus-specific glycoprotein S13 precursor, putative [Ricinus communis] |
| 7 |
Hb_003754_080 |
0.2468978284 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 8 |
Hb_009644_040 |
0.2491936613 |
- |
- |
PREDICTED: protein SKIP34 [Jatropha curcas] |
| 9 |
Hb_000316_020 |
0.2524478364 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 10 |
Hb_081504_010 |
0.2634231848 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |
| 11 |
Hb_108528_010 |
0.2649834187 |
- |
- |
unnamed protein product [Coffea canephora] |
| 12 |
Hb_001150_030 |
0.2671179795 |
- |
- |
NADP-dependent oxidoreductase P1 isoform 3 [Theobroma cacao] |
| 13 |
Hb_003052_210 |
0.27091839 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002151_050 |
0.2730087136 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 15 |
Hb_000115_050 |
0.273038362 |
- |
- |
- |
| 16 |
Hb_029920_040 |
0.2738455533 |
- |
- |
PREDICTED: uncharacterized protein LOC105635837 [Jatropha curcas] |
| 17 |
Hb_000084_040 |
0.2746255311 |
- |
- |
hypothetical protein RCOM_1007940 [Ricinus communis] |
| 18 |
Hb_179520_010 |
0.275678584 |
- |
- |
PREDICTED: uncharacterized protein LOC105763691 [Gossypium raimondii] |
| 19 |
Hb_001842_130 |
0.2765645841 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002943_030 |
0.2793069821 |
- |
- |
- |