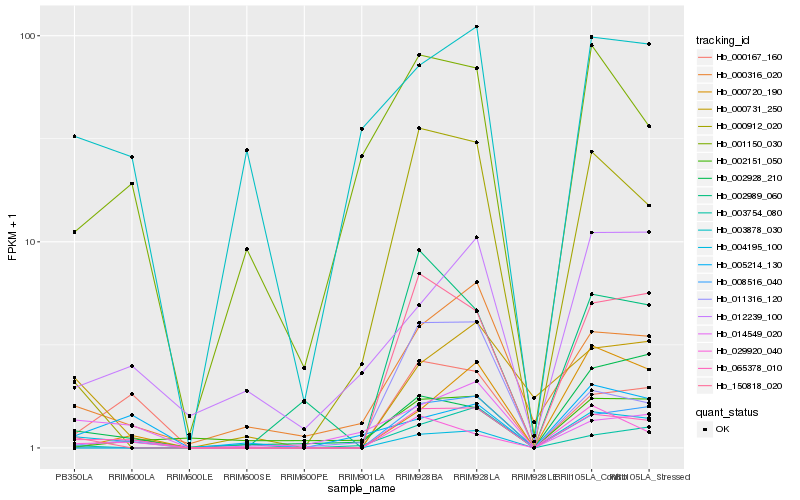
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_065378_010 |
0.0 |
- |
- |
S-locus-specific glycoprotein S13 precursor, putative [Ricinus communis] |
| 2 |
Hb_000167_160 |
0.1658125953 |
- |
- |
- |
| 3 |
Hb_000912_020 |
0.2099600618 |
- |
- |
PREDICTED: uncharacterized protein LOC105645039 [Jatropha curcas] |
| 4 |
Hb_150818_020 |
0.2168102688 |
- |
- |
PREDICTED: uncharacterized protein LOC105645039 [Jatropha curcas] |
| 5 |
Hb_029920_040 |
0.2206764529 |
- |
- |
PREDICTED: uncharacterized protein LOC105635837 [Jatropha curcas] |
| 6 |
Hb_000316_020 |
0.2241746306 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 7 |
Hb_008516_040 |
0.2297047104 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004195_100 |
0.2386188926 |
- |
- |
PREDICTED: uncharacterized protein LOC104106427 [Nicotiana tomentosiformis] |
| 9 |
Hb_001150_030 |
0.2528683628 |
- |
- |
NADP-dependent oxidoreductase P1 isoform 3 [Theobroma cacao] |
| 10 |
Hb_011316_120 |
0.2610679747 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH118-like [Jatropha curcas] |
| 11 |
Hb_000731_250 |
0.2685033226 |
- |
- |
- |
| 12 |
Hb_014549_020 |
0.2695821843 |
- |
- |
hypothetical protein POPTR_0013s12350g [Populus trichocarpa] |
| 13 |
Hb_012239_100 |
0.2732546465 |
- |
- |
hypothetical protein VITISV_043897 [Vitis vinifera] |
| 14 |
Hb_000720_190 |
0.2737859494 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 15 |
Hb_005214_130 |
0.2779368219 |
- |
- |
- |
| 16 |
Hb_002151_050 |
0.2784321311 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 17 |
Hb_003878_030 |
0.2792563822 |
- |
- |
hypothetical protein POPTR_0011s11120g [Populus trichocarpa] |
| 18 |
Hb_002989_060 |
0.2806256034 |
- |
- |
hypothetical protein JCGZ_22461 [Jatropha curcas] |
| 19 |
Hb_002928_210 |
0.2828001468 |
- |
- |
- |
| 20 |
Hb_003754_080 |
0.2849352929 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |