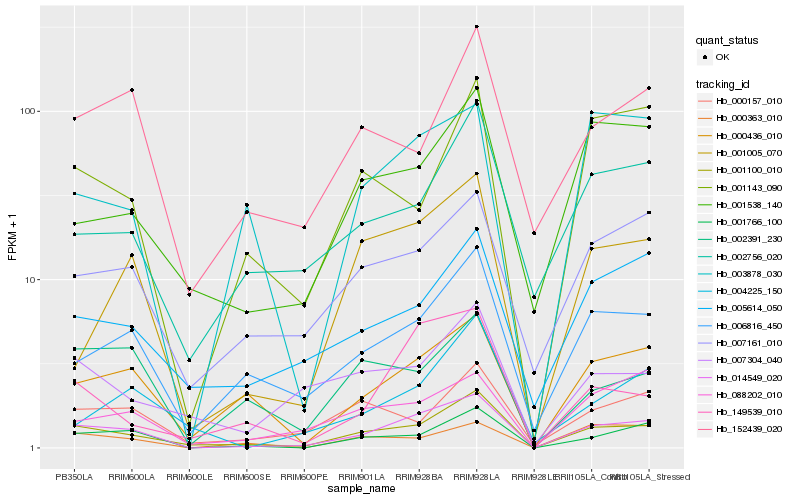
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_014549_020 |
0.0 |
- |
- |
hypothetical protein POPTR_0013s12350g [Populus trichocarpa] |
| 2 |
Hb_001100_010 |
0.1326642845 |
- |
- |
F-box/FBD/LRR-repeat protein [Medicago truncatula] |
| 3 |
Hb_001766_100 |
0.1630127786 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_000436_010 |
0.1776850249 |
- |
- |
PREDICTED: probable polyamine transporter At1g31830 isoform X1 [Eucalyptus grandis] |
| 5 |
Hb_006816_450 |
0.180101266 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_007161_010 |
0.1959677061 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105630404 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001005_070 |
0.2006361748 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF113-like [Jatropha curcas] |
| 8 |
Hb_005614_050 |
0.2089467068 |
- |
- |
PREDICTED: aspartate aminotransferase, mitochondrial-like [Jatropha curcas] |
| 9 |
Hb_088202_010 |
0.2095584987 |
- |
- |
hypothetical protein JCGZ_13241 [Jatropha curcas] |
| 10 |
Hb_149539_010 |
0.2114322219 |
- |
- |
- |
| 11 |
Hb_003878_030 |
0.2193161064 |
- |
- |
hypothetical protein POPTR_0011s11120g [Populus trichocarpa] |
| 12 |
Hb_000157_010 |
0.2197338876 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 13 |
Hb_001538_140 |
0.219990074 |
- |
- |
PREDICTED: PXMP2/4 family protein 4-like [Jatropha curcas] |
| 14 |
Hb_002756_020 |
0.2200436076 |
transcription factor |
TF Family: HB |
bel1 homeotic protein, putative [Ricinus communis] |
| 15 |
Hb_002391_230 |
0.2226059377 |
- |
- |
hypothetical protein RCOM_0745080 [Ricinus communis] |
| 16 |
Hb_152439_020 |
0.2238179597 |
- |
- |
PREDICTED: copper-transporting ATPase RAN1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_007304_040 |
0.224254235 |
- |
- |
PREDICTED: polyadenylate-binding protein 3 [Jatropha curcas] |
| 18 |
Hb_001143_090 |
0.2245114977 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 6 [Jatropha curcas] |
| 19 |
Hb_000363_010 |
0.2278683573 |
- |
- |
- |
| 20 |
Hb_004225_150 |
0.2309857584 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger protein 36, C3H1 type-like 3 [Jatropha curcas] |