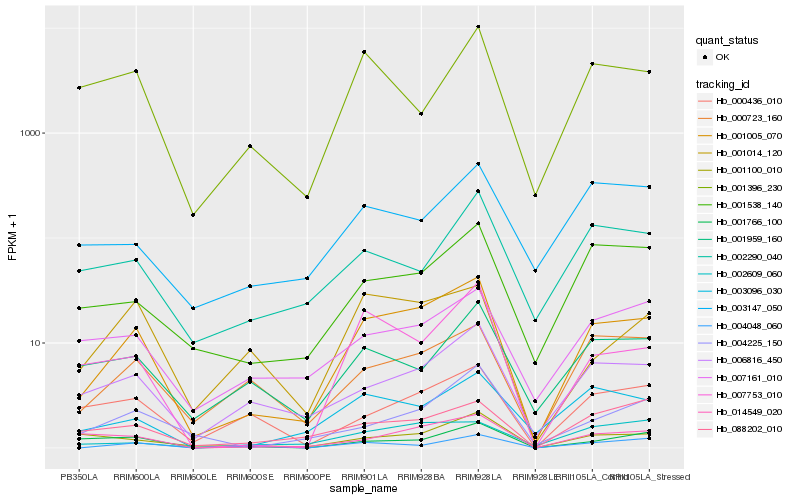
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001005_070 |
0.0 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF113-like [Jatropha curcas] |
| 2 |
Hb_006816_450 |
0.1819882087 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003096_030 |
0.1952618424 |
- |
- |
PREDICTED: bifunctional riboflavin biosynthesis protein RIBA 1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000723_160 |
0.1966689119 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP11 [Jatropha curcas] |
| 5 |
Hb_001538_140 |
0.2000682671 |
- |
- |
PREDICTED: PXMP2/4 family protein 4-like [Jatropha curcas] |
| 6 |
Hb_014549_020 |
0.2006361748 |
- |
- |
hypothetical protein POPTR_0013s12350g [Populus trichocarpa] |
| 7 |
Hb_007753_010 |
0.2039633187 |
- |
- |
- |
| 8 |
Hb_001014_120 |
0.2116443822 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor ERF113 [Jatropha curcas] |
| 9 |
Hb_004225_150 |
0.2138592117 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger protein 36, C3H1 type-like 3 [Jatropha curcas] |
| 10 |
Hb_088202_010 |
0.2142202796 |
- |
- |
hypothetical protein JCGZ_13241 [Jatropha curcas] |
| 11 |
Hb_007161_010 |
0.2167425457 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105630404 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001100_010 |
0.2168957793 |
- |
- |
F-box/FBD/LRR-repeat protein [Medicago truncatula] |
| 13 |
Hb_002609_060 |
0.218030011 |
- |
- |
PREDICTED: sugar transport protein 8-like [Jatropha curcas] |
| 14 |
Hb_000436_010 |
0.2203549419 |
- |
- |
PREDICTED: probable polyamine transporter At1g31830 isoform X1 [Eucalyptus grandis] |
| 15 |
Hb_001959_160 |
0.2256149567 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g12770-like [Jatropha curcas] |
| 16 |
Hb_004048_060 |
0.2279174455 |
- |
- |
hypothetical protein PRUPE_ppa023788mg, partial [Prunus persica] |
| 17 |
Hb_003147_050 |
0.2282004355 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 18 |
Hb_002290_040 |
0.2286045379 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001396_230 |
0.2291251856 |
- |
- |
hypothetical protein MIMGU_mgv1a023992mg, partial [Erythranthe guttata] |
| 20 |
Hb_001766_100 |
0.2297905752 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |