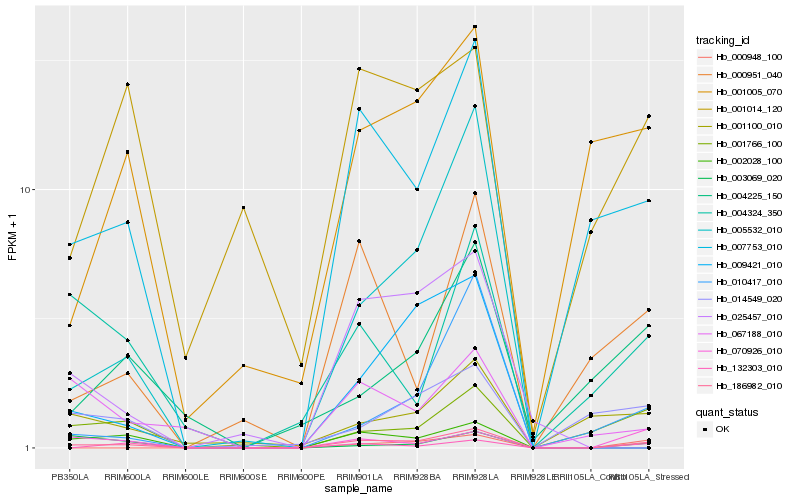
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_186982_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104120222 [Nicotiana tomentosiformis] |
| 2 |
Hb_007753_010 |
0.2121430002 |
- |
- |
- |
| 3 |
Hb_132303_010 |
0.2777110531 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 4 |
Hb_070926_010 |
0.2793661083 |
- |
- |
PREDICTED: mRNA-decapping enzyme subunit 2-like [Populus euphratica] |
| 5 |
Hb_001766_100 |
0.279999849 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_000951_040 |
0.2846718034 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_09931 [Jatropha curcas] |
| 7 |
Hb_000948_100 |
0.2928810377 |
- |
- |
- |
| 8 |
Hb_010417_010 |
0.2941357589 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 9 |
Hb_001005_070 |
0.294669337 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF113-like [Jatropha curcas] |
| 10 |
Hb_001100_010 |
0.2973049103 |
- |
- |
F-box/FBD/LRR-repeat protein [Medicago truncatula] |
| 11 |
Hb_002028_100 |
0.298417117 |
- |
- |
PREDICTED: uncharacterized protein LOC102624085 [Citrus sinensis] |
| 12 |
Hb_067188_010 |
0.2998831872 |
- |
- |
- |
| 13 |
Hb_014549_020 |
0.3021673446 |
- |
- |
hypothetical protein POPTR_0013s12350g [Populus trichocarpa] |
| 14 |
Hb_004324_350 |
0.302517196 |
- |
- |
PREDICTED: uncharacterized protein LOC103484146 [Cucumis melo] |
| 15 |
Hb_003069_020 |
0.3026470736 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004225_150 |
0.3078357724 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger protein 36, C3H1 type-like 3 [Jatropha curcas] |
| 17 |
Hb_025457_010 |
0.3111810558 |
- |
- |
- |
| 18 |
Hb_001014_120 |
0.3117915967 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor ERF113 [Jatropha curcas] |
| 19 |
Hb_005532_010 |
0.3118798969 |
- |
- |
- |
| 20 |
Hb_009421_010 |
0.3134469055 |
- |
- |
- |