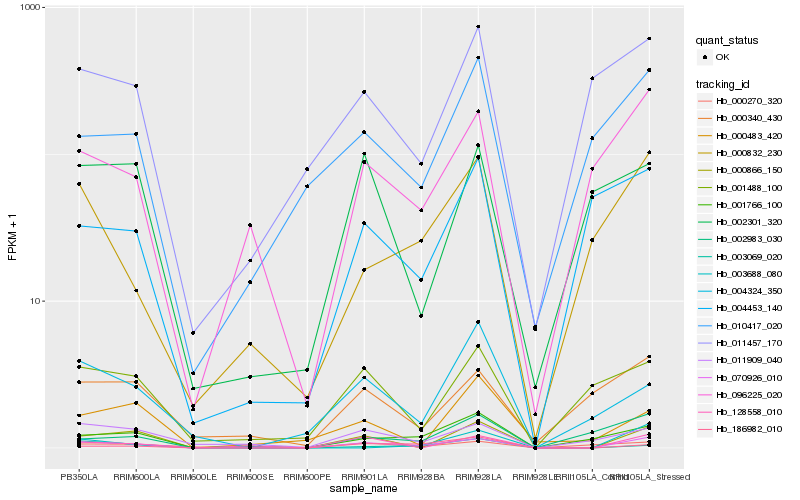
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_070926_010 |
0.0 |
- |
- |
PREDICTED: mRNA-decapping enzyme subunit 2-like [Populus euphratica] |
| 2 |
Hb_000866_150 |
0.2145568558 |
- |
- |
- |
| 3 |
Hb_128558_010 |
0.2504875512 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 4 |
Hb_001766_100 |
0.261048156 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_003069_020 |
0.2658710356 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_096225_020 |
0.2664519783 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT22-like [Jatropha curcas] |
| 7 |
Hb_011909_040 |
0.275639554 |
- |
- |
hypothetical protein JCGZ_00750 [Jatropha curcas] |
| 8 |
Hb_000270_320 |
0.2772467473 |
- |
- |
hypothetical protein RCOM_1512920 [Ricinus communis] |
| 9 |
Hb_000832_230 |
0.2792868337 |
transcription factor |
TF Family: C2H2 |
hypothetical protein JCGZ_18434 [Jatropha curcas] |
| 10 |
Hb_186982_010 |
0.2793661083 |
- |
- |
PREDICTED: uncharacterized protein LOC104120222 [Nicotiana tomentosiformis] |
| 11 |
Hb_004324_350 |
0.2839914901 |
- |
- |
PREDICTED: uncharacterized protein LOC103484146 [Cucumis melo] |
| 12 |
Hb_001488_100 |
0.2893376806 |
- |
- |
- |
| 13 |
Hb_010417_020 |
0.2904299257 |
- |
- |
interferon-induced GTP-binding protein mx, putative [Ricinus communis] |
| 14 |
Hb_000340_430 |
0.2910745748 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_011457_170 |
0.2968761646 |
- |
- |
PREDICTED: chlorophyllase-1-like [Jatropha curcas] |
| 16 |
Hb_002983_030 |
0.2971594032 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein CISIN_1g046124mg, partial [Citrus sinensis] |
| 17 |
Hb_003688_080 |
0.3010455807 |
- |
- |
- |
| 18 |
Hb_004453_140 |
0.3034392097 |
- |
- |
PREDICTED: uncharacterized protein LOC104880181 [Vitis vinifera] |
| 19 |
Hb_002301_320 |
0.3049965702 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0004s04590g [Populus trichocarpa] |
| 20 |
Hb_000483_420 |
0.308598578 |
- |
- |
PREDICTED: putative glycerol-3-phosphate transporter 4 [Jatropha curcas] |