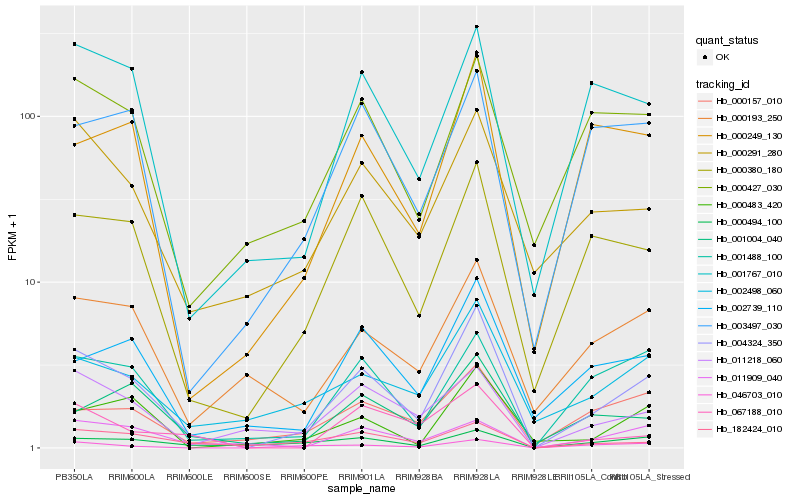
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004324_350 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103484146 [Cucumis melo] |
| 2 |
Hb_000380_180 |
0.1748911087 |
- |
- |
kinase, putative [Ricinus communis] |
| 3 |
Hb_001488_100 |
0.1966474046 |
- |
- |
- |
| 4 |
Hb_001767_010 |
0.1979068053 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Populus euphratica] |
| 5 |
Hb_002498_060 |
0.1981558107 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_002739_110 |
0.1987645358 |
- |
- |
PREDICTED: ribosomal RNA processing protein 1 homolog [Jatropha curcas] |
| 7 |
Hb_000249_130 |
0.1992281469 |
- |
- |
hypothetical protein RCOM_0254640 [Ricinus communis] |
| 8 |
Hb_000291_280 |
0.2015536896 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: myosin-11 [Jatropha curcas] |
| 9 |
Hb_011218_060 |
0.2036583668 |
- |
- |
PREDICTED: dephospho-CoA kinase-like [Jatropha curcas] |
| 10 |
Hb_000157_010 |
0.2055259885 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 11 |
Hb_001004_040 |
0.206292062 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_182424_010 |
0.2073180212 |
- |
- |
PREDICTED: U-box domain-containing protein 51 [Jatropha curcas] |
| 13 |
Hb_000193_250 |
0.2097722751 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g61400 [Jatropha curcas] |
| 14 |
Hb_000427_030 |
0.2111567729 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000483_420 |
0.2115109687 |
- |
- |
PREDICTED: putative glycerol-3-phosphate transporter 4 [Jatropha curcas] |
| 16 |
Hb_003497_030 |
0.2122607588 |
- |
- |
ADP-ribosylation factor, putative [Ricinus communis] |
| 17 |
Hb_046703_010 |
0.2165565665 |
- |
- |
Leucine-rich repeat protein kinase family protein [Theobroma cacao] |
| 18 |
Hb_011909_040 |
0.2174913472 |
- |
- |
hypothetical protein JCGZ_00750 [Jatropha curcas] |
| 19 |
Hb_067188_010 |
0.2196213169 |
- |
- |
- |
| 20 |
Hb_000494_100 |
0.2233005048 |
- |
- |
H(\+)-transporting atpase plant/fungi plasma membrane type, putative [Ricinus communis] |