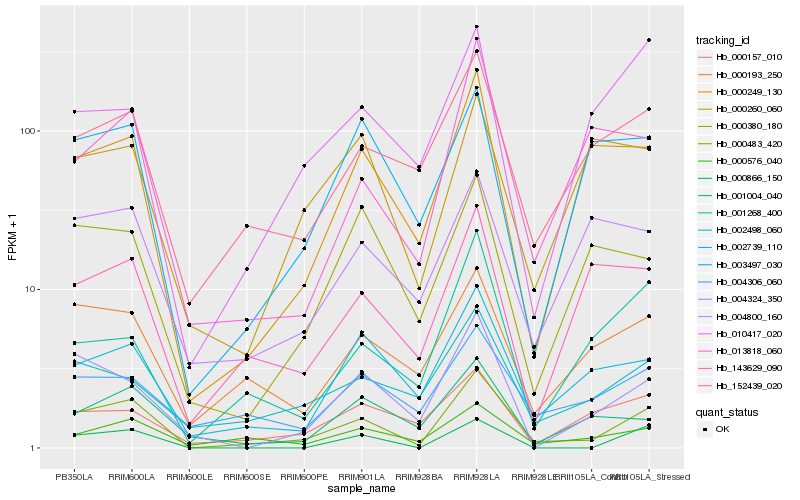
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000483_420 |
0.0 |
- |
- |
PREDICTED: putative glycerol-3-phosphate transporter 4 [Jatropha curcas] |
| 2 |
Hb_000576_040 |
0.1993120829 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g15130 [Jatropha curcas] |
| 3 |
Hb_002739_110 |
0.2043379849 |
- |
- |
PREDICTED: ribosomal RNA processing protein 1 homolog [Jatropha curcas] |
| 4 |
Hb_152439_020 |
0.2051030141 |
- |
- |
PREDICTED: copper-transporting ATPase RAN1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000193_250 |
0.2081169115 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g61400 [Jatropha curcas] |
| 6 |
Hb_004324_350 |
0.2115109687 |
- |
- |
PREDICTED: uncharacterized protein LOC103484146 [Cucumis melo] |
| 7 |
Hb_000249_130 |
0.2180659117 |
- |
- |
hypothetical protein RCOM_0254640 [Ricinus communis] |
| 8 |
Hb_002498_060 |
0.2187185173 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_013818_060 |
0.2192520436 |
- |
- |
Brassinosteroid LRR receptor kinase precursor, putative [Ricinus communis] |
| 10 |
Hb_004306_060 |
0.2212610084 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g25060, mitochondrial [Jatropha curcas] |
| 11 |
Hb_001268_400 |
0.2243312294 |
- |
- |
PREDICTED: uncharacterized protein LOC104583970 [Brachypodium distachyon] |
| 12 |
Hb_143629_090 |
0.2254386678 |
- |
- |
PREDICTED: uncharacterized protein LOC105647358 [Jatropha curcas] |
| 13 |
Hb_000866_150 |
0.2271362915 |
- |
- |
- |
| 14 |
Hb_001004_040 |
0.2305523076 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003497_030 |
0.2326189967 |
- |
- |
ADP-ribosylation factor, putative [Ricinus communis] |
| 16 |
Hb_000260_060 |
0.2344798364 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 6 [Jatropha curcas] |
| 17 |
Hb_010417_020 |
0.2380097161 |
- |
- |
interferon-induced GTP-binding protein mx, putative [Ricinus communis] |
| 18 |
Hb_000380_180 |
0.2381416939 |
- |
- |
kinase, putative [Ricinus communis] |
| 19 |
Hb_004800_160 |
0.2383117818 |
- |
- |
Uncharacterized protein TCM_000403 [Theobroma cacao] |
| 20 |
Hb_000157_010 |
0.2384017295 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |