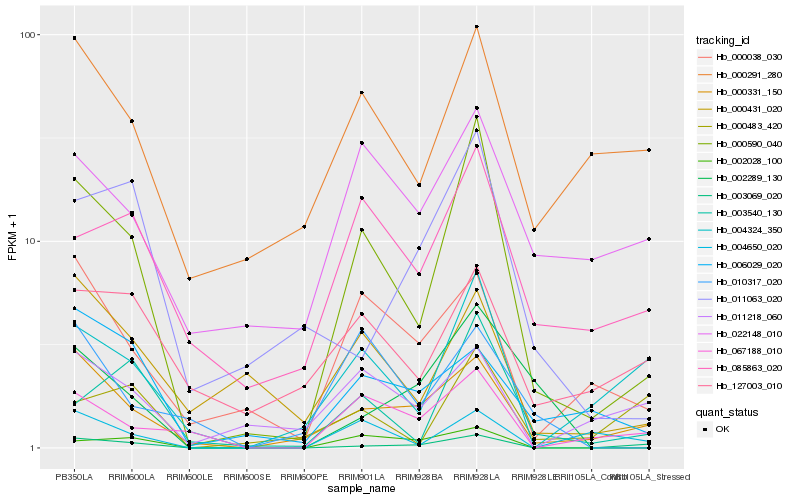
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000590_040 |
0.0 |
- |
- |
hypothetical protein MPER_09217 [Moniliophthora perniciosa FA553] |
| 2 |
Hb_003540_130 |
0.2324418288 |
- |
- |
- |
| 3 |
Hb_000331_150 |
0.2374507161 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 4 |
Hb_003069_020 |
0.2465276639 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004324_350 |
0.2471072554 |
- |
- |
PREDICTED: uncharacterized protein LOC103484146 [Cucumis melo] |
| 6 |
Hb_002028_100 |
0.2622456144 |
- |
- |
PREDICTED: uncharacterized protein LOC102624085 [Citrus sinensis] |
| 7 |
Hb_067188_010 |
0.2646132007 |
- |
- |
- |
| 8 |
Hb_000038_030 |
0.2674964481 |
- |
- |
PREDICTED: probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 7-like [Glycine max] |
| 9 |
Hb_085863_020 |
0.2692031016 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 1 [Jatropha curcas] |
| 10 |
Hb_000431_020 |
0.2725594133 |
- |
- |
hypothetical protein JCGZ_06801 [Jatropha curcas] |
| 11 |
Hb_004650_020 |
0.2805081331 |
- |
- |
- |
| 12 |
Hb_000291_280 |
0.2823269876 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: myosin-11 [Jatropha curcas] |
| 13 |
Hb_002289_130 |
0.2876612487 |
transcription factor |
TF Family: C2H2 |
arsenite inducuble RNA associated protein aip-1, putative [Ricinus communis] |
| 14 |
Hb_011063_020 |
0.2932933037 |
- |
- |
PREDICTED: ribosomal L1 domain-containing protein 1-like [Jatropha curcas] |
| 15 |
Hb_010317_020 |
0.2955149837 |
- |
- |
PREDICTED: sarcoplasmic reticulum histidine-rich calcium-binding protein [Jatropha curcas] |
| 16 |
Hb_022148_010 |
0.2973530165 |
- |
- |
PREDICTED: uncharacterized protein LOC105638910 [Jatropha curcas] |
| 17 |
Hb_011218_060 |
0.2993898466 |
- |
- |
PREDICTED: dephospho-CoA kinase-like [Jatropha curcas] |
| 18 |
Hb_006029_020 |
0.2994098621 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1, partial [Vitis vinifera] |
| 19 |
Hb_000483_420 |
0.2999304632 |
- |
- |
PREDICTED: putative glycerol-3-phosphate transporter 4 [Jatropha curcas] |
| 20 |
Hb_127003_010 |
0.3012343077 |
- |
- |
PREDICTED: uncharacterized protein LOC105633613 [Jatropha curcas] |