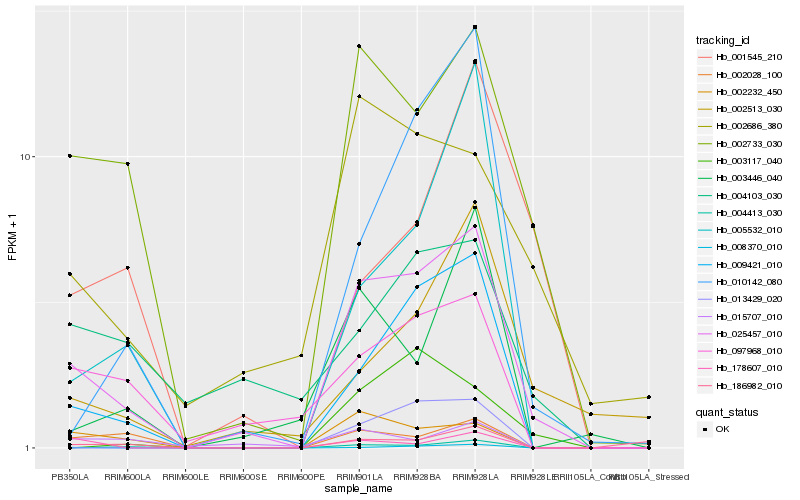
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_025457_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_002733_030 |
0.1926758637 |
- |
- |
PREDICTED: uncharacterized protein LOC104248159 [Nicotiana sylvestris] |
| 3 |
Hb_009421_010 |
0.2036683764 |
- |
- |
- |
| 4 |
Hb_097968_010 |
0.2479667244 |
- |
- |
hypothetical protein PRUPE_ppa001143mg [Prunus persica] |
| 5 |
Hb_002232_450 |
0.2486933372 |
- |
- |
PREDICTED: probable methyltransferase PMT11 [Jatropha curcas] |
| 6 |
Hb_002686_380 |
0.2568406975 |
- |
- |
- |
| 7 |
Hb_008370_010 |
0.2705731845 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 8 |
Hb_002028_100 |
0.2716243965 |
- |
- |
PREDICTED: uncharacterized protein LOC102624085 [Citrus sinensis] |
| 9 |
Hb_010142_080 |
0.2746817109 |
- |
- |
PREDICTED: uncharacterized protein LOC105630660 [Jatropha curcas] |
| 10 |
Hb_004413_030 |
0.2782944786 |
- |
- |
Retrotransposon protein, putative [Theobroma cacao] |
| 11 |
Hb_004103_030 |
0.2788604841 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002513_030 |
0.2864466971 |
- |
- |
PREDICTED: alpha-L-fucosidase 2-like isoform X2 [Elaeis guineensis] |
| 13 |
Hb_003446_040 |
0.2886125263 |
- |
- |
- |
| 14 |
Hb_013429_020 |
0.2915192011 |
- |
- |
prolyl 4-hydroxylase alpha subunit, putative [Ricinus communis] |
| 15 |
Hb_005532_010 |
0.2929661819 |
- |
- |
- |
| 16 |
Hb_015707_010 |
0.293449204 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 17 |
Hb_003117_040 |
0.2989738978 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 18 |
Hb_001545_210 |
0.3008849538 |
- |
- |
hypothetical protein JCGZ_11132 [Jatropha curcas] |
| 19 |
Hb_178607_010 |
0.304511399 |
- |
- |
PREDICTED: uncharacterized protein LOC105789535 [Gossypium raimondii] |
| 20 |
Hb_186982_010 |
0.3111810558 |
- |
- |
PREDICTED: uncharacterized protein LOC104120222 [Nicotiana tomentosiformis] |