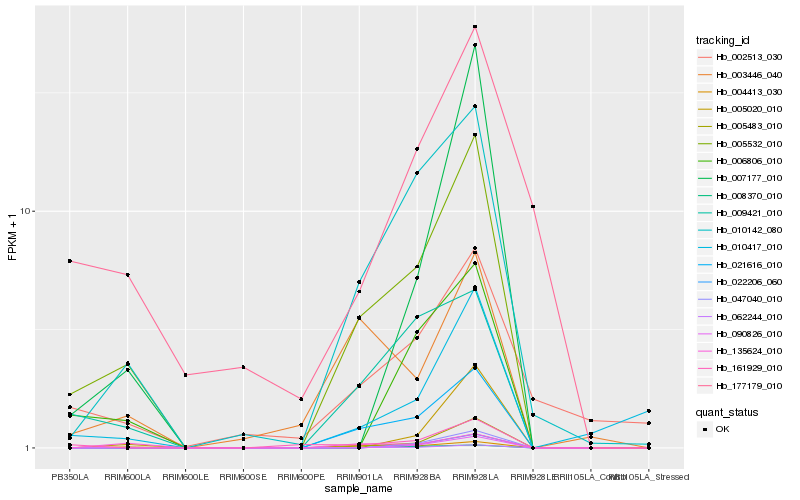
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005532_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_062244_010 |
0.1621083336 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 3 |
Hb_010142_080 |
0.1774384204 |
- |
- |
PREDICTED: uncharacterized protein LOC105630660 [Jatropha curcas] |
| 4 |
Hb_021616_010 |
0.1900370029 |
- |
- |
- |
| 5 |
Hb_009421_010 |
0.2010122488 |
- |
- |
- |
| 6 |
Hb_004413_030 |
0.2046205423 |
- |
- |
Retrotransposon protein, putative [Theobroma cacao] |
| 7 |
Hb_005483_010 |
0.2158544705 |
- |
- |
PREDICTED: replication protein A 70 kDa DNA-binding subunit A [Jatropha curcas] |
| 8 |
Hb_008370_010 |
0.2260187824 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 9 |
Hb_135624_010 |
0.235360991 |
- |
- |
PREDICTED: inositol transporter 4-like [Jatropha curcas] |
| 10 |
Hb_006806_010 |
0.2376130751 |
- |
- |
- |
| 11 |
Hb_161929_010 |
0.2523787091 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 12 |
Hb_003446_040 |
0.2533430611 |
- |
- |
- |
| 13 |
Hb_010417_010 |
0.261256654 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 14 |
Hb_007177_010 |
0.2687994612 |
- |
- |
- |
| 15 |
Hb_005020_010 |
0.2717413989 |
- |
- |
- |
| 16 |
Hb_177179_010 |
0.2744268917 |
- |
- |
PREDICTED: putative 4-hydroxy-4-methyl-2-oxoglutarate aldolase 2 [Populus euphratica] |
| 17 |
Hb_002513_030 |
0.2755806593 |
- |
- |
PREDICTED: alpha-L-fucosidase 2-like isoform X2 [Elaeis guineensis] |
| 18 |
Hb_022206_060 |
0.284829665 |
- |
- |
- |
| 19 |
Hb_047040_010 |
0.2848477971 |
- |
- |
integrase [Gossypium herbaceum] |
| 20 |
Hb_090826_010 |
0.2849094834 |
- |
- |
PREDICTED: uncharacterized protein LOC105786910 [Gossypium raimondii] |