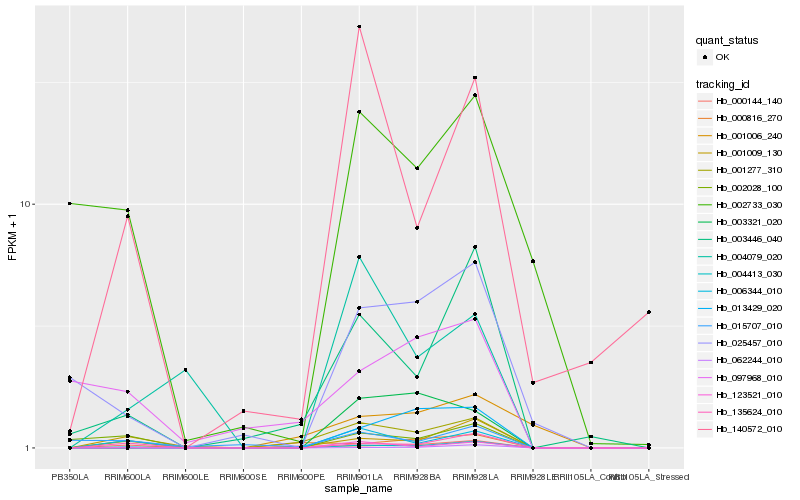
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001277_310 |
0.0 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Phoenix dactylifera] |
| 2 |
Hb_004413_030 |
0.2209399601 |
- |
- |
Retrotransposon protein, putative [Theobroma cacao] |
| 3 |
Hb_000144_140 |
0.222187812 |
- |
- |
PREDICTED: uncharacterized protein LOC105629608 [Jatropha curcas] |
| 4 |
Hb_003446_040 |
0.2335388349 |
- |
- |
- |
| 5 |
Hb_006344_010 |
0.2413573693 |
- |
- |
- |
| 6 |
Hb_001009_130 |
0.2621250031 |
- |
- |
hypothetical protein JCGZ_11336 [Jatropha curcas] |
| 7 |
Hb_135624_010 |
0.2816324444 |
- |
- |
PREDICTED: inositol transporter 4-like [Jatropha curcas] |
| 8 |
Hb_001006_240 |
0.285760396 |
- |
- |
- |
| 9 |
Hb_140572_010 |
0.2979515954 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 7-like [Jatropha curcas] |
| 10 |
Hb_123521_010 |
0.3054056706 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 11 |
Hb_025457_010 |
0.3213760973 |
- |
- |
- |
| 12 |
Hb_097968_010 |
0.3273638853 |
- |
- |
hypothetical protein PRUPE_ppa001143mg [Prunus persica] |
| 13 |
Hb_003321_020 |
0.3341254434 |
- |
- |
PREDICTED: TMV resistance protein N-like, partial [Citrus sinensis] |
| 14 |
Hb_013429_020 |
0.3355709984 |
- |
- |
prolyl 4-hydroxylase alpha subunit, putative [Ricinus communis] |
| 15 |
Hb_002028_100 |
0.3356790589 |
- |
- |
PREDICTED: uncharacterized protein LOC102624085 [Citrus sinensis] |
| 16 |
Hb_000816_270 |
0.3367741731 |
- |
- |
- |
| 17 |
Hb_015707_010 |
0.3383619754 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 18 |
Hb_004079_020 |
0.3395616739 |
- |
- |
hypothetical protein glysoja_031528 [Glycine soja] |
| 19 |
Hb_062244_010 |
0.3398852722 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 20 |
Hb_002733_030 |
0.342786084 |
- |
- |
PREDICTED: uncharacterized protein LOC104248159 [Nicotiana sylvestris] |