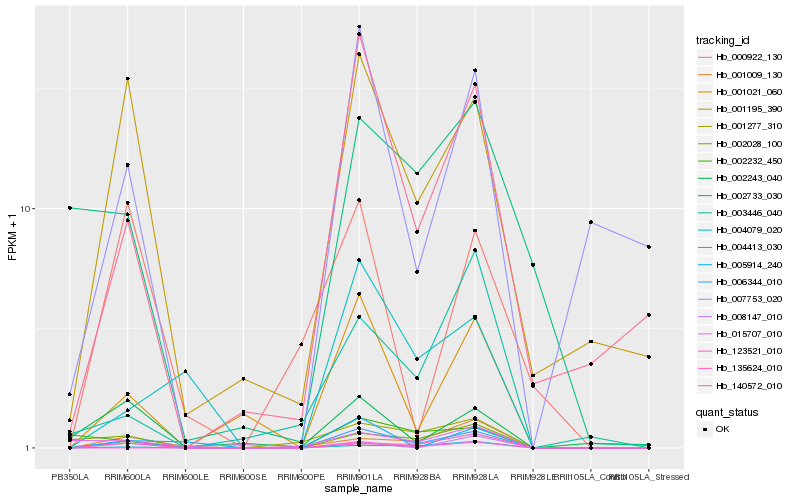
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006344_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_140572_010 |
0.1989134524 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 7-like [Jatropha curcas] |
| 3 |
Hb_001021_060 |
0.2138817366 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_001195_390 |
0.229376593 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 7-like [Populus euphratica] |
| 5 |
Hb_004413_030 |
0.2331842246 |
- |
- |
Retrotransposon protein, putative [Theobroma cacao] |
| 6 |
Hb_001277_310 |
0.2413573693 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Phoenix dactylifera] |
| 7 |
Hb_135624_010 |
0.2535404402 |
- |
- |
PREDICTED: inositol transporter 4-like [Jatropha curcas] |
| 8 |
Hb_007753_020 |
0.2623595539 |
- |
- |
- |
| 9 |
Hb_123521_010 |
0.2775348019 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 10 |
Hb_001009_130 |
0.2793405132 |
- |
- |
hypothetical protein JCGZ_11336 [Jatropha curcas] |
| 11 |
Hb_008147_010 |
0.2865443774 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 12 |
Hb_002028_100 |
0.291936817 |
- |
- |
PREDICTED: uncharacterized protein LOC102624085 [Citrus sinensis] |
| 13 |
Hb_004079_020 |
0.2935930712 |
- |
- |
hypothetical protein glysoja_031528 [Glycine soja] |
| 14 |
Hb_003446_040 |
0.2952388366 |
- |
- |
- |
| 15 |
Hb_005914_240 |
0.3036701826 |
- |
- |
PREDICTED: oxidation resistance protein 1 [Jatropha curcas] |
| 16 |
Hb_002232_450 |
0.3198949502 |
- |
- |
PREDICTED: probable methyltransferase PMT11 [Jatropha curcas] |
| 17 |
Hb_002243_040 |
0.3243822137 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_00227 [Jatropha curcas] |
| 18 |
Hb_002733_030 |
0.3252165519 |
- |
- |
PREDICTED: uncharacterized protein LOC104248159 [Nicotiana sylvestris] |
| 19 |
Hb_015707_010 |
0.3438368611 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 20 |
Hb_000922_130 |
0.3447540429 |
- |
- |
hypothetical protein POPTR_0008s04925g [Populus trichocarpa] |