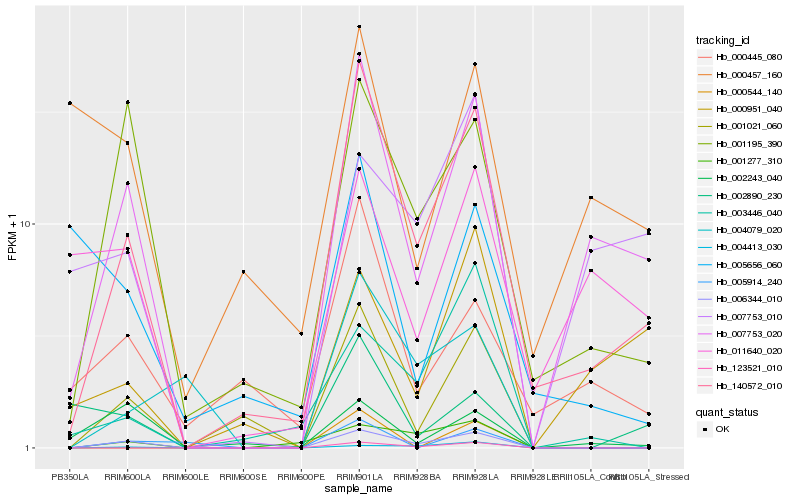
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_140572_010 |
0.0 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 7-like [Jatropha curcas] |
| 2 |
Hb_007753_020 |
0.1770803051 |
- |
- |
- |
| 3 |
Hb_006344_010 |
0.1989134524 |
- |
- |
- |
| 4 |
Hb_001021_060 |
0.2300372188 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_001195_390 |
0.2487041385 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 7-like [Populus euphratica] |
| 6 |
Hb_000445_080 |
0.2571399917 |
transcription factor |
TF Family: MIKC |
PREDICTED: floral homeotic protein PMADS 2 [Jatropha curcas] |
| 7 |
Hb_123521_010 |
0.2749303068 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 8 |
Hb_001277_310 |
0.2979515954 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Phoenix dactylifera] |
| 9 |
Hb_004079_020 |
0.3035969637 |
- |
- |
hypothetical protein glysoja_031528 [Glycine soja] |
| 10 |
Hb_003446_040 |
0.3057932491 |
- |
- |
- |
| 11 |
Hb_000951_040 |
0.3091678263 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_09931 [Jatropha curcas] |
| 12 |
Hb_002890_230 |
0.3131238605 |
- |
- |
hypothetical protein MTR_5g084920 [Medicago truncatula] |
| 13 |
Hb_011640_020 |
0.3236146251 |
- |
- |
hypothetical protein VITISV_036432 [Vitis vinifera] |
| 14 |
Hb_005914_240 |
0.3236627007 |
- |
- |
PREDICTED: oxidation resistance protein 1 [Jatropha curcas] |
| 15 |
Hb_000544_140 |
0.3246822165 |
- |
- |
PREDICTED: uncharacterized protein LOC101245866 [Solanum lycopersicum] |
| 16 |
Hb_004413_030 |
0.329985276 |
- |
- |
Retrotransposon protein, putative [Theobroma cacao] |
| 17 |
Hb_007753_010 |
0.3327495456 |
- |
- |
- |
| 18 |
Hb_002243_040 |
0.333270453 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_00227 [Jatropha curcas] |
| 19 |
Hb_000457_160 |
0.3390400702 |
- |
- |
PREDICTED: uncharacterized protein LOC105633076 [Jatropha curcas] |
| 20 |
Hb_005656_060 |
0.3391521171 |
- |
- |
- |