| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
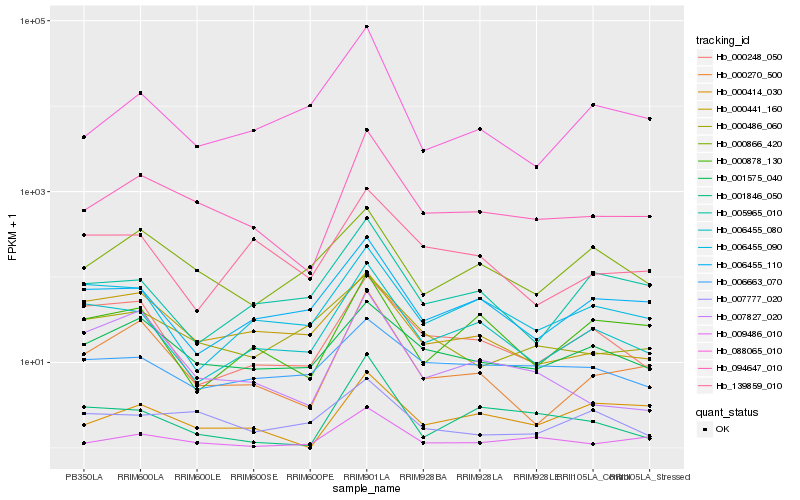
Hb_094647_010 |
0.0 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 5-B-like [Jatropha curcas] |
| 2 |
Hb_000270_500 |
0.1544627027 |
- |
- |
PREDICTED: uncharacterized protein LOC105640439 [Jatropha curcas] |
| 3 |
Hb_009486_010 |
0.1623694941 |
- |
- |
- |
| 4 |
Hb_001575_040 |
0.1923658856 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 25 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000414_030 |
0.2010325582 |
- |
- |
- |
| 6 |
Hb_006455_080 |
0.2036885172 |
- |
- |
PREDICTED: transcription factor GTE6-like isoform X2 [Nelumbo nucifera] |
| 7 |
Hb_006455_110 |
0.205439478 |
- |
- |
bromodomain-containing protein, putative [Ricinus communis] |
| 8 |
Hb_000486_060 |
0.2069967069 |
- |
- |
PREDICTED: immediate early response 3-interacting protein 1-like isoform X2 [Glycine max] |
| 9 |
Hb_000866_420 |
0.2078323944 |
- |
- |
Remorin, putative [Ricinus communis] |
| 10 |
Hb_007827_020 |
0.2085527926 |
- |
- |
- |
| 11 |
Hb_005965_010 |
0.2173199489 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb family transcription factor APL-like [Jatropha curcas] |
| 12 |
Hb_006455_090 |
0.2182597267 |
- |
- |
PREDICTED: transcription factor GTE1-like [Jatropha curcas] |
| 13 |
Hb_088065_010 |
0.2186141636 |
- |
- |
hypothetical protein glysoja_007934 [Glycine soja] |
| 14 |
Hb_001846_050 |
0.2204075039 |
- |
- |
- |
| 15 |
Hb_000441_160 |
0.2210613746 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 16 |
Hb_000248_050 |
0.2215024177 |
- |
- |
PREDICTED: uncharacterized protein LOC105647198 isoform X3 [Jatropha curcas] |
| 17 |
Hb_139859_010 |
0.2225250912 |
- |
- |
PREDICTED: cell division control protein 48 homolog D-like [Nicotiana sylvestris] |
| 18 |
Hb_006663_070 |
0.223970963 |
- |
- |
PREDICTED: uncharacterized protein LOC105637432 [Jatropha curcas] |
| 19 |
Hb_007777_020 |
0.2248617128 |
- |
- |
hypothetical protein JCGZ_14410 [Jatropha curcas] |
| 20 |
Hb_000878_130 |
0.2253863792 |
- |
- |
conserved hypothetical protein [Ricinus communis] |