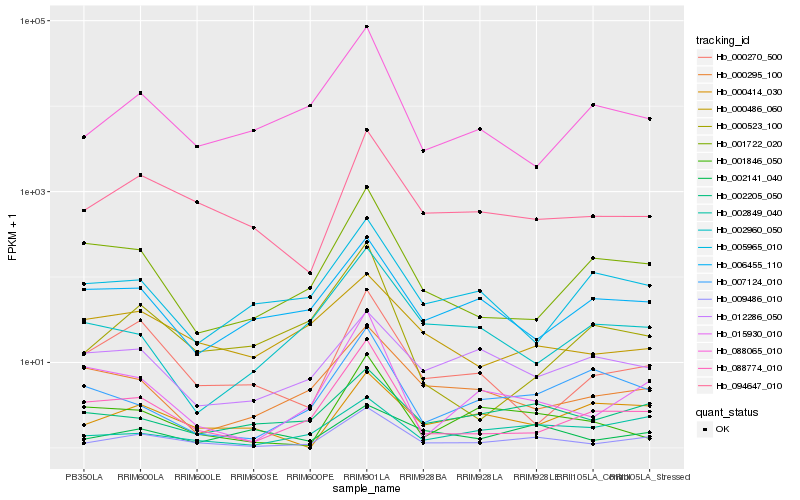
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009486_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_094647_010 |
0.1623694941 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 5-B-like [Jatropha curcas] |
| 3 |
Hb_002205_050 |
0.2040979645 |
- |
- |
hypothetical protein JCGZ_03335 [Jatropha curcas] |
| 4 |
Hb_088774_010 |
0.2112075421 |
- |
- |
- |
| 5 |
Hb_002849_040 |
0.2136441381 |
- |
- |
caax prenyl protease ste24, putative [Ricinus communis] |
| 6 |
Hb_015930_010 |
0.2173685801 |
- |
- |
uncharacterized protein LOC100499930 [Glycine max] |
| 7 |
Hb_088065_010 |
0.2225386934 |
- |
- |
hypothetical protein glysoja_007934 [Glycine soja] |
| 8 |
Hb_000270_500 |
0.232476712 |
- |
- |
PREDICTED: uncharacterized protein LOC105640439 [Jatropha curcas] |
| 9 |
Hb_000486_060 |
0.2340819113 |
- |
- |
PREDICTED: immediate early response 3-interacting protein 1-like isoform X2 [Glycine max] |
| 10 |
Hb_006455_110 |
0.2388373575 |
- |
- |
bromodomain-containing protein, putative [Ricinus communis] |
| 11 |
Hb_000295_100 |
0.2395963982 |
- |
- |
- |
| 12 |
Hb_002141_040 |
0.2396324347 |
- |
- |
- |
| 13 |
Hb_001722_020 |
0.2408380944 |
- |
- |
- |
| 14 |
Hb_001846_050 |
0.2431932308 |
- |
- |
- |
| 15 |
Hb_002960_050 |
0.2472510379 |
- |
- |
PREDICTED: uncharacterized protein LOC105635325 [Jatropha curcas] |
| 16 |
Hb_005965_010 |
0.2499043797 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb family transcription factor APL-like [Jatropha curcas] |
| 17 |
Hb_000414_030 |
0.2510507462 |
- |
- |
- |
| 18 |
Hb_007124_010 |
0.2522215442 |
- |
- |
unnamed protein product [Coffea canephora] |
| 19 |
Hb_012286_050 |
0.2543016171 |
- |
- |
PREDICTED: syntaxin-61 [Jatropha curcas] |
| 20 |
Hb_000523_100 |
0.255037376 |
- |
- |
PREDICTED: protein DJ-1 homolog D-like [Populus euphratica] |