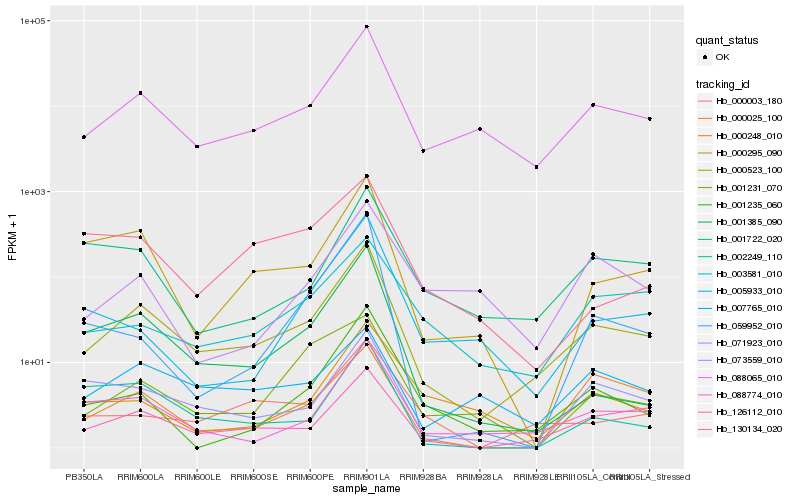
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000523_100 |
0.0 |
- |
- |
PREDICTED: protein DJ-1 homolog D-like [Populus euphratica] |
| 2 |
Hb_088065_010 |
0.119892891 |
- |
- |
hypothetical protein glysoja_007934 [Glycine soja] |
| 3 |
Hb_126112_010 |
0.1563304045 |
- |
- |
PREDICTED: uncharacterized protein LOC105636715 [Jatropha curcas] |
| 4 |
Hb_088774_010 |
0.1630879599 |
- |
- |
- |
| 5 |
Hb_000295_090 |
0.1827973026 |
- |
- |
Clathrin light chain 2 [Glycine soja] |
| 6 |
Hb_000003_180 |
0.1856235687 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_071923_010 |
0.1973633641 |
- |
- |
- |
| 8 |
Hb_001235_060 |
0.1974317458 |
- |
- |
- |
| 9 |
Hb_001385_090 |
0.1994202744 |
- |
- |
PREDICTED: uncharacterized protein LOC105644404 isoform X2 [Jatropha curcas] |
| 10 |
Hb_003581_010 |
0.2071773114 |
- |
- |
hypothetical protein VITISV_004687 [Vitis vinifera] |
| 11 |
Hb_001722_020 |
0.2071789835 |
- |
- |
- |
| 12 |
Hb_000248_010 |
0.2116452349 |
- |
- |
hypothetical protein PRUPE_ppa008056mg [Prunus persica] |
| 13 |
Hb_002249_110 |
0.2191313377 |
- |
- |
PREDICTED: centromere protein S isoform X1 [Jatropha curcas] |
| 14 |
Hb_007765_010 |
0.2204083104 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_005933_010 |
0.2206952495 |
- |
- |
PREDICTED: putative disease resistance RPP13-like protein 1 [Populus euphratica] |
| 16 |
Hb_000025_100 |
0.2233795029 |
- |
- |
- |
| 17 |
Hb_073559_010 |
0.224094254 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 12 [Jatropha curcas] |
| 18 |
Hb_001231_070 |
0.2277568649 |
- |
- |
- |
| 19 |
Hb_130134_020 |
0.2320104848 |
- |
- |
- |
| 20 |
Hb_059952_010 |
0.2353942177 |
- |
- |
PREDICTED: probable protein S-acyltransferase 7 [Eucalyptus grandis] |