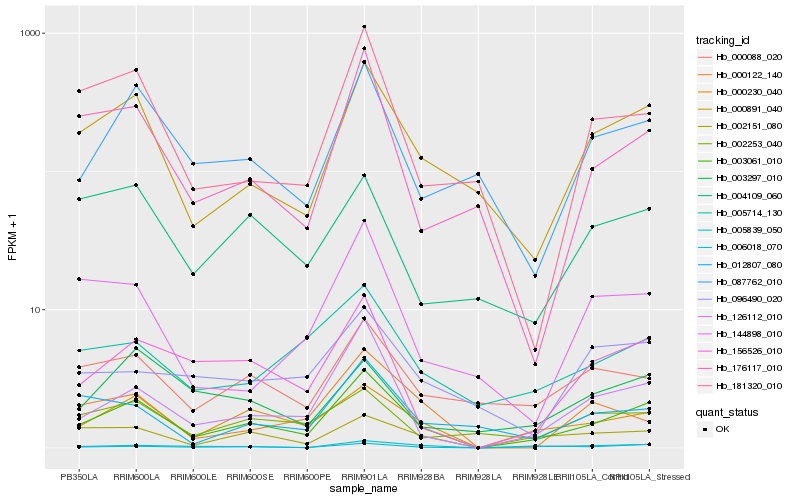
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003061_010 |
0.0 |
- |
- |
hypothetical protein SORBIDRAFT_01g039870 [Sorghum bicolor] |
| 2 |
Hb_000230_040 |
0.1503601674 |
- |
- |
PREDICTED: protein MON2 homolog isoform X2 [Vitis vinifera] |
| 3 |
Hb_126112_010 |
0.1636011731 |
- |
- |
PREDICTED: uncharacterized protein LOC105636715 [Jatropha curcas] |
| 4 |
Hb_000891_040 |
0.1655983429 |
- |
- |
hypothetical protein JCGZ_11252 [Jatropha curcas] |
| 5 |
Hb_156526_010 |
0.1737215897 |
- |
- |
PREDICTED: uncharacterized protein LOC105636715 [Jatropha curcas] |
| 6 |
Hb_002151_080 |
0.194569782 |
- |
- |
hypothetical protein MIMGU_mgv1a000572mg [Erythranthe guttata] |
| 7 |
Hb_005839_050 |
0.1950203672 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 8 |
Hb_087762_010 |
0.1985180369 |
- |
- |
thioredoxin-like family protein [Hevea brasiliensis] |
| 9 |
Hb_005714_130 |
0.2069034846 |
transcription factor |
TF Family: C2H2 |
unknown [Populus trichocarpa x Populus deltoides] |
| 10 |
Hb_000088_020 |
0.2069992887 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_10946 [Jatropha curcas] |
| 11 |
Hb_176117_010 |
0.2099181084 |
- |
- |
Cold-inducible RNA-binding protein [Morus notabilis] |
| 12 |
Hb_006018_070 |
0.2114391015 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 13 |
Hb_181320_010 |
0.2155712124 |
- |
- |
PREDICTED: signal recognition particle 14 kDa protein [Jatropha curcas] |
| 14 |
Hb_002253_040 |
0.2180526728 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g13690 [Jatropha curcas] |
| 15 |
Hb_012807_080 |
0.2228301593 |
- |
- |
hypothetical protein CICLE_v10028099mg [Citrus clementina] |
| 16 |
Hb_004109_060 |
0.2232064316 |
- |
- |
hypothetical protein POPTR_0019s12520g [Populus trichocarpa] |
| 17 |
Hb_144898_010 |
0.2274965083 |
- |
- |
PREDICTED: 60S ribosomal protein L7-2-like [Jatropha curcas] |
| 18 |
Hb_003297_010 |
0.2298829292 |
- |
- |
PREDICTED: uncharacterized protein LOC105636715 [Jatropha curcas] |
| 19 |
Hb_000122_140 |
0.2299604605 |
- |
- |
- |
| 20 |
Hb_096490_020 |
0.2304008723 |
- |
- |
PREDICTED: UPF0565 protein C2orf69 [Jatropha curcas] |