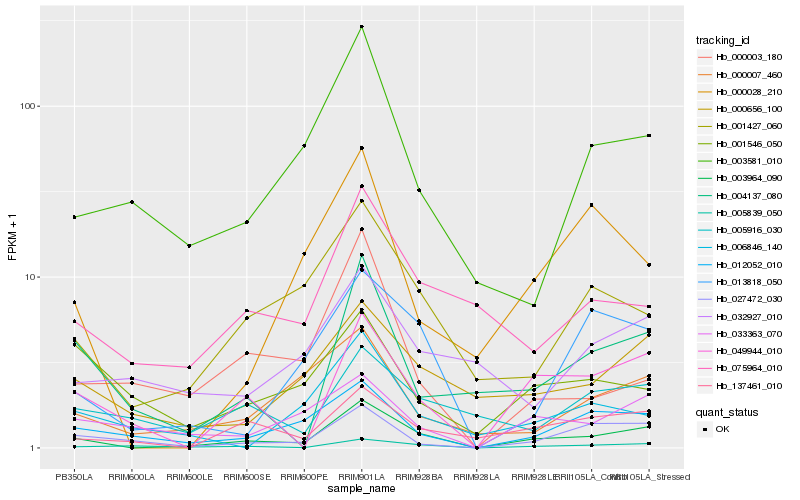
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003964_090 |
0.0 |
- |
- |
PREDICTED: 14-3-3-like protein GF14 iota [Jatropha curcas] |
| 2 |
Hb_005839_050 |
0.2291107753 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 3 |
Hb_049944_010 |
0.22931189 |
- |
- |
transducin family protein [Populus trichocarpa] |
| 4 |
Hb_012052_010 |
0.2393503622 |
- |
- |
PREDICTED: uncharacterized protein LOC103323653 [Prunus mume] |
| 5 |
Hb_001427_060 |
0.240444575 |
- |
- |
- |
| 6 |
Hb_013818_050 |
0.2449582166 |
- |
- |
PREDICTED: uncharacterized protein LOC105629143 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000656_100 |
0.2459690377 |
- |
- |
PREDICTED: uncharacterized protein LOC105633651 [Jatropha curcas] |
| 8 |
Hb_137461_010 |
0.2475211051 |
- |
- |
hypothetical protein POPTR_0011s03680g [Populus trichocarpa] |
| 9 |
Hb_027472_030 |
0.2514879195 |
- |
- |
RNA helicase 36, partial [Manihot esculenta] |
| 10 |
Hb_033363_070 |
0.25893928 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: methionine aminopeptidase 1B, chloroplastic, partial [Populus euphratica] |
| 11 |
Hb_000007_460 |
0.2624029597 |
- |
- |
PREDICTED: uncharacterized protein LOC105641948 [Jatropha curcas] |
| 12 |
Hb_075964_010 |
0.2646064034 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 13 |
Hb_004137_080 |
0.2653822934 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003581_010 |
0.2685579724 |
- |
- |
hypothetical protein VITISV_004687 [Vitis vinifera] |
| 15 |
Hb_005916_030 |
0.2761917209 |
- |
- |
PREDICTED: uncharacterized protein At2g29880-like [Jatropha curcas] |
| 16 |
Hb_000028_210 |
0.2785596302 |
- |
- |
hypothetical protein CISIN_1g0103302mg, partial [Citrus sinensis] |
| 17 |
Hb_032927_010 |
0.2914831603 |
- |
- |
- |
| 18 |
Hb_006846_140 |
0.2920691253 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000003_180 |
0.2930495313 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001546_050 |
0.2968184475 |
- |
- |
- |