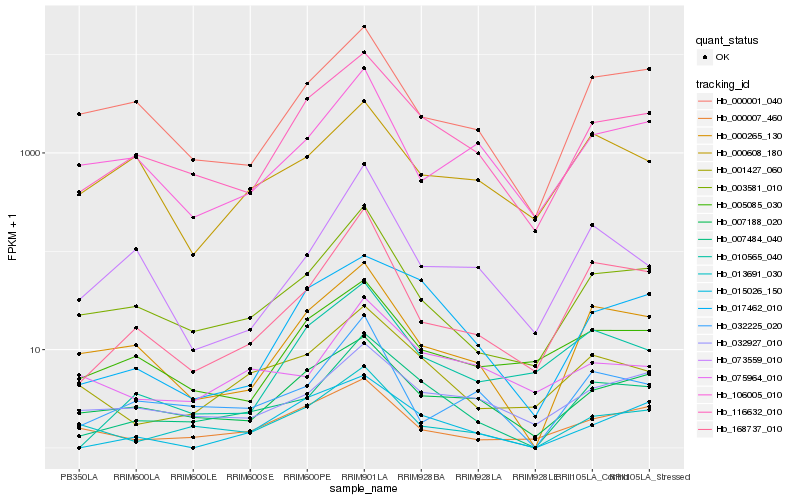
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007484_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_116632_010 |
0.151801165 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_003581_010 |
0.1641801213 |
- |
- |
hypothetical protein VITISV_004687 [Vitis vinifera] |
| 4 |
Hb_168737_010 |
0.1840682139 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_001427_060 |
0.1872428247 |
- |
- |
- |
| 6 |
Hb_000265_130 |
0.1882011616 |
- |
- |
- |
| 7 |
Hb_013691_030 |
0.1929839492 |
- |
- |
hypothetical protein CISIN_1g020131mg [Citrus sinensis] |
| 8 |
Hb_017462_010 |
0.1969622205 |
- |
- |
- |
| 9 |
Hb_007188_020 |
0.2012036492 |
- |
- |
hypothetical protein PRUPE_ppb010436mg [Prunus persica] |
| 10 |
Hb_032927_010 |
0.2046512554 |
- |
- |
- |
| 11 |
Hb_032225_020 |
0.2051287387 |
- |
- |
hypothetical protein POPTR_0006s08140g [Populus trichocarpa] |
| 12 |
Hb_000001_040 |
0.207117724 |
- |
- |
unnamed protein product [Coffea canephora] |
| 13 |
Hb_075964_010 |
0.2147730644 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 14 |
Hb_073559_010 |
0.2267754939 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 12 [Jatropha curcas] |
| 15 |
Hb_000608_180 |
0.230331027 |
- |
- |
PREDICTED: putative 4-hydroxy-4-methyl-2-oxoglutarate aldolase 2 [Tarenaya hassleriana] |
| 16 |
Hb_000007_460 |
0.2319338098 |
- |
- |
PREDICTED: uncharacterized protein LOC105641948 [Jatropha curcas] |
| 17 |
Hb_106005_010 |
0.2324496716 |
- |
- |
EG5651 [Manihot esculenta] |
| 18 |
Hb_010565_040 |
0.2334682845 |
- |
- |
hypothetical protein POPTR_0015s05280g, partial [Populus trichocarpa] |
| 19 |
Hb_015026_150 |
0.2343456292 |
- |
- |
Nuclease PA3, putative [Ricinus communis] |
| 20 |
Hb_005085_030 |
0.23459889 |
- |
- |
PREDICTED: N-acetyltransferase 9-like protein isoform X1 [Jatropha curcas] |