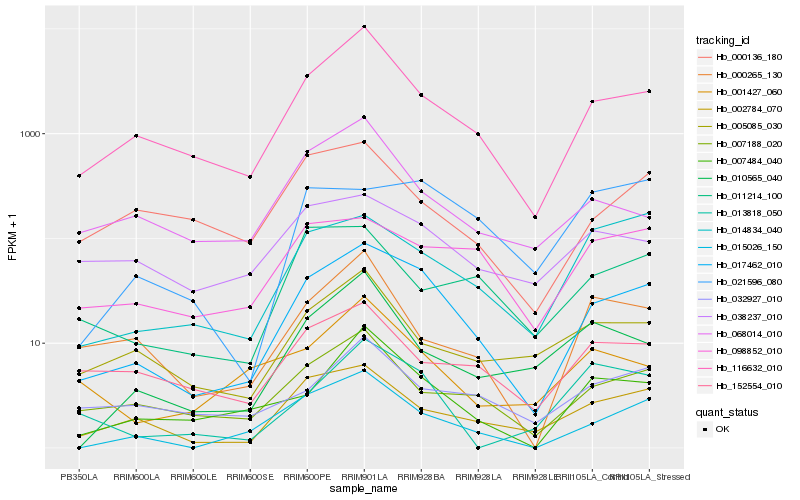
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_017462_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_116632_010 |
0.1496353612 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_015026_150 |
0.1817101013 |
- |
- |
Nuclease PA3, putative [Ricinus communis] |
| 4 |
Hb_002784_070 |
0.1829398559 |
- |
- |
BnaUnng01470D [Brassica napus] |
| 5 |
Hb_007188_020 |
0.1953753509 |
- |
- |
hypothetical protein PRUPE_ppb010436mg [Prunus persica] |
| 6 |
Hb_007484_040 |
0.1969622205 |
- |
- |
- |
| 7 |
Hb_014834_040 |
0.2027133031 |
- |
- |
hypothetical protein JCGZ_03409 [Jatropha curcas] |
| 8 |
Hb_098852_010 |
0.2096632282 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 9 |
Hb_005085_030 |
0.2261710526 |
- |
- |
PREDICTED: N-acetyltransferase 9-like protein isoform X1 [Jatropha curcas] |
| 10 |
Hb_152554_010 |
0.2267917346 |
- |
- |
hypothetical protein B456_008G095800, partial [Gossypium raimondii] |
| 11 |
Hb_021596_080 |
0.2269865241 |
- |
- |
50S ribosomal protein L33A [Hevea brasiliensis] |
| 12 |
Hb_011214_100 |
0.2275664412 |
- |
- |
PREDICTED: uncharacterized protein LOC105633003 [Jatropha curcas] |
| 13 |
Hb_038237_010 |
0.2281518963 |
- |
- |
hypothetical protein POPTR_0003s01110g [Populus trichocarpa] |
| 14 |
Hb_000265_130 |
0.2283948303 |
- |
- |
- |
| 15 |
Hb_032927_010 |
0.2292412831 |
- |
- |
- |
| 16 |
Hb_001427_060 |
0.2308012592 |
- |
- |
- |
| 17 |
Hb_000136_180 |
0.2310119255 |
- |
- |
ribosomal protein S28, putative [Ricinus communis] |
| 18 |
Hb_013818_050 |
0.233842779 |
- |
- |
PREDICTED: uncharacterized protein LOC105629143 isoform X2 [Jatropha curcas] |
| 19 |
Hb_010565_040 |
0.2354358417 |
- |
- |
hypothetical protein POPTR_0015s05280g, partial [Populus trichocarpa] |
| 20 |
Hb_068014_010 |
0.2369085711 |
- |
- |
hypothetical protein POPTR_0312s00200g, partial [Populus trichocarpa] |