| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
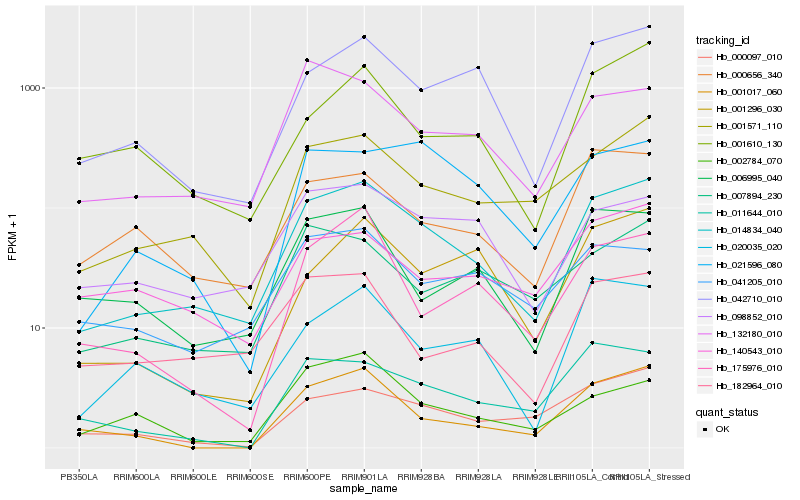
Hb_014834_040 |
0.0 |
- |
- |
hypothetical protein JCGZ_03409 [Jatropha curcas] |
| 2 |
Hb_001571_110 |
0.1252374374 |
- |
- |
PREDICTED: 60S ribosomal protein L44-like [Gossypium raimondii] |
| 3 |
Hb_042710_010 |
0.1419366709 |
- |
- |
PREDICTED: 40S ribosomal protein S15a-1 [Jatropha curcas] |
| 4 |
Hb_098852_010 |
0.1494864963 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 5 |
Hb_132180_010 |
0.1576224288 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 6 |
Hb_020035_020 |
0.1579415124 |
- |
- |
PREDICTED: uncharacterized protein LOC105631020 [Jatropha curcas] |
| 7 |
Hb_006995_040 |
0.1582423106 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: 14 kDa zinc-binding protein-like [Pyrus x bretschneideri] |
| 8 |
Hb_000656_340 |
0.1590882797 |
- |
- |
hypothetical protein JCGZ_24686 [Jatropha curcas] |
| 9 |
Hb_182964_010 |
0.1601089564 |
- |
- |
PREDICTED: uncharacterized protein LOC105642044 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000097_010 |
0.1626940004 |
- |
- |
- |
| 11 |
Hb_001017_060 |
0.1629129268 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 12 |
Hb_007894_230 |
0.1660328988 |
- |
- |
30S ribosomal protein S14, putative [Ricinus communis] |
| 13 |
Hb_021596_080 |
0.1661266789 |
- |
- |
50S ribosomal protein L33A [Hevea brasiliensis] |
| 14 |
Hb_011644_010 |
0.1677826064 |
- |
- |
PREDICTED: probable choline-phosphate cytidylyltransferase [Elaeis guineensis] |
| 15 |
Hb_041205_010 |
0.172712846 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_175976_010 |
0.1736101023 |
- |
- |
- |
| 17 |
Hb_001610_130 |
0.1753276499 |
- |
- |
40S ribosomal protein S30 [Medicago truncatula] |
| 18 |
Hb_140543_010 |
0.1754767213 |
- |
- |
- |
| 19 |
Hb_002784_070 |
0.178546089 |
- |
- |
BnaUnng01470D [Brassica napus] |
| 20 |
Hb_001296_030 |
0.1792225399 |
- |
- |
PREDICTED: uncharacterized protein LOC105638276 [Jatropha curcas] |