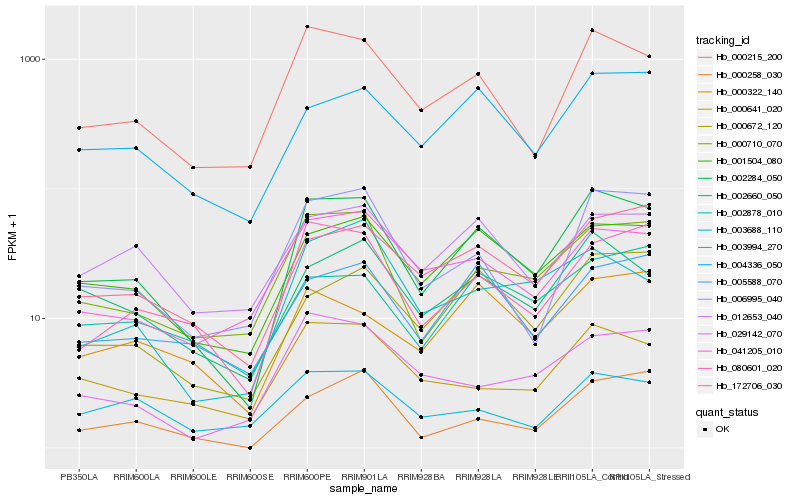
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002284_050 |
0.0 |
- |
- |
hypothetical protein POPTR_0002s00740g [Populus trichocarpa] |
| 2 |
Hb_000215_200 |
0.1118979502 |
- |
- |
RecName: Full=Profilin-2; AltName: Full=Pollen allergen Hev b 8.0102; AltName: Allergen=Hev b 8.0102 [Hevea brasiliensis] |
| 3 |
Hb_001504_080 |
0.1210700268 |
- |
- |
golgi snare 11 protein, putative [Ricinus communis] |
| 4 |
Hb_000710_070 |
0.1263942164 |
- |
- |
PREDICTED: uncharacterized protein LOC105645553 [Jatropha curcas] |
| 5 |
Hb_080601_020 |
0.1315492558 |
- |
- |
PREDICTED: cytochrome c oxidase copper chaperone 1 [Jatropha curcas] |
| 6 |
Hb_002660_050 |
0.1358716141 |
- |
- |
PREDICTED: uncharacterized protein LOC105642218 [Jatropha curcas] |
| 7 |
Hb_172706_030 |
0.1367576018 |
- |
- |
PREDICTED: uncharacterized protein LOC105640907 [Jatropha curcas] |
| 8 |
Hb_006995_040 |
0.1370285693 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: 14 kDa zinc-binding protein-like [Pyrus x bretschneideri] |
| 9 |
Hb_004336_050 |
0.1417816939 |
- |
- |
hypothetical protein JCGZ_26651 [Jatropha curcas] |
| 10 |
Hb_000672_120 |
0.1426455956 |
- |
- |
V-type proton ATPase subunit G 1 [Jatropha curcas] |
| 11 |
Hb_000641_020 |
0.1486226165 |
- |
- |
PREDICTED: tubulin-folding cofactor B-like [Jatropha curcas] |
| 12 |
Hb_041205_010 |
0.1487269968 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_005588_070 |
0.1533559359 |
- |
- |
hypothetical protein JCGZ_04298 [Jatropha curcas] |
| 14 |
Hb_012653_040 |
0.1552880063 |
- |
- |
UV-induced protein uvi31, putative [Ricinus communis] |
| 15 |
Hb_000258_030 |
0.1620083656 |
- |
- |
- |
| 16 |
Hb_002878_010 |
0.1623843086 |
- |
- |
- |
| 17 |
Hb_000322_140 |
0.1635480485 |
- |
- |
hypothetical protein RCOM_1047200 [Ricinus communis] |
| 18 |
Hb_003994_270 |
0.1642979167 |
- |
- |
PREDICTED: uncharacterized protein LOC105646152 [Jatropha curcas] |
| 19 |
Hb_003688_110 |
0.164961189 |
- |
- |
- |
| 20 |
Hb_029142_070 |
0.1682250381 |
- |
- |
- |