Hb_000215_200
Information
| Type | - |
|---|---|
| Description | - |
| Location | Contig215: 167410-169723 |
| Sequence |   |
Annotation
kegg
| ID | tcc:TCM_037757 |
|---|---|
| description | Profilin 1 isoform 1 |
nr
| ID | Q9STB6.1 |
|---|---|
| description | RecName: Full=Profilin-2; AltName: Full=Pollen allergen Hev b 8.0102; AltName: Allergen=Hev b 8.0102 [Hevea brasiliensis] |
swissprot
| ID | Q9STB6 |
|---|---|
| description | Profilin-2 OS=Hevea brasiliensis GN=PRO2 PE=1 SV=1 |
trembl
| ID | I3SDB5 |
|---|---|
| description | Profilin OS=Lotus japonicus PE=2 SV=1 |
Gene Ontology
| ID | GO:0005737 |
|---|---|
| description | profilin 1 isoform 1 |
Full-length cDNA clone information
| cDNA+EST (Sanger&Illumina) (ID:Location) |
PASA_asmbl_22194: 167763-169843 |
|---|---|
| cDNA (Sanger) (ID:Location) |
005_F07.ab1: 167918-169802 , 005_I17.ab1: 167846-169727 , 008_F04.ab1: 167846-169727 , 009_J05.ab1: 167864-169727 , 015_F05.ab1: 167785-169727 , 016_G16.ab1: 167855-169657 , 020_A03.ab1: 167890-169805 , 021_O01.ab1: 167818-169725 , 025_N10.ab1: 167826-169749 , 028_D07.ab1: 168082-169721 , 028_L20.ab1: 167815-169727 , 030_B09.ab1: 167869-169749 , 030_L20.ab1: 167909-169794 , 031_G04.ab1: 167824-169707 , 033_H02.ab1: 167823-169727 , 034_C22.ab1: 167890-169794 , 035_I09.ab1: 167913-169727 , 035_J08.ab1: 167890-169843 , 036_A07.ab1: 167832-169725 , 040_D21.ab1: 167961-169806 , 047_K04.ab1: 167911-169806 , 051_P10.ab1: 167864-169719 , 052_N05.ab1: 167867-167951 |
Similar expressed genes (Top20)
Gene co-expression network
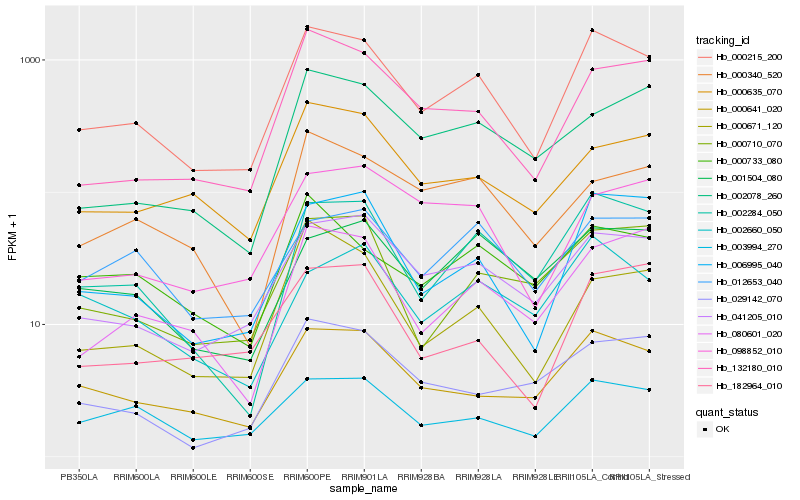
Expression pattern by RNA-Seq analysis
| RRIM600_Latex | RRIM600_Bark | RRIM600_Leaf | RRIM600_Petiole | PB350_Latex | RRIM901_Latex |
|---|---|---|---|---|---|
| 331.731 | 147.043 | 144.82 | 1786.29 | 294.762 | 1406.91 |
| RRII105_Latex_C | RRII105_Latex_S | RRIM928_Latex | RRIM928_Bark | RRIM928_Leaf | |
| 1675.77 | 1050.06 | 770.14 | 402.521 | 175.694 |

