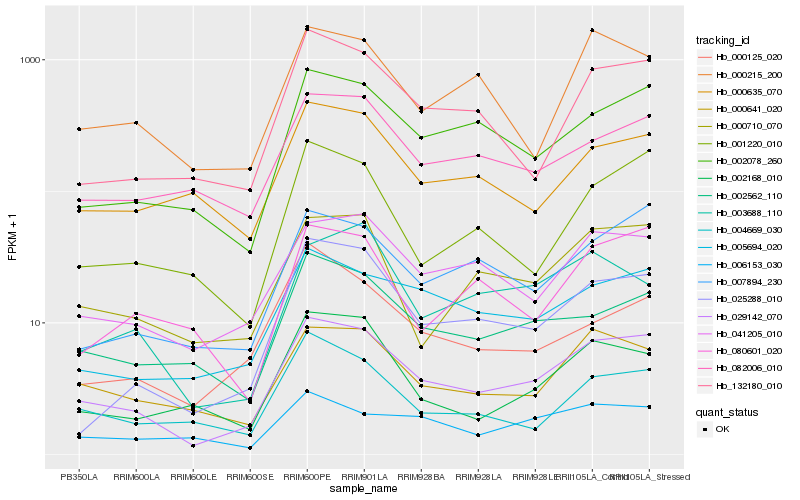
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029142_070 |
0.0 |
- |
- |
- |
| 2 |
Hb_041205_010 |
0.1252695083 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002078_260 |
0.1258089729 |
- |
- |
unknown [Populus trichocarpa] |
| 4 |
Hb_005694_020 |
0.1279090027 |
- |
- |
PREDICTED: ORM1-like protein 2 [Gossypium raimondii] |
| 5 |
Hb_000641_020 |
0.1294493187 |
- |
- |
PREDICTED: tubulin-folding cofactor B-like [Jatropha curcas] |
| 6 |
Hb_132180_010 |
0.134465965 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 7 |
Hb_007894_230 |
0.1350761763 |
- |
- |
30S ribosomal protein S14, putative [Ricinus communis] |
| 8 |
Hb_000710_070 |
0.1365699727 |
- |
- |
PREDICTED: uncharacterized protein LOC105645553 [Jatropha curcas] |
| 9 |
Hb_025288_010 |
0.1392103829 |
- |
- |
PREDICTED: protein unc-50 homolog [Jatropha curcas] |
| 10 |
Hb_002562_110 |
0.1401389934 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX3, mitochondrial [Jatropha curcas] |
| 11 |
Hb_082006_010 |
0.1477551354 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001220_010 |
0.1511785509 |
- |
- |
hypothetical protein JCGZ_00411 [Jatropha curcas] |
| 13 |
Hb_002168_010 |
0.1530703823 |
- |
- |
PREDICTED: bifunctional dihydrofolate reductase-thymidylate synthase-like isoform X4 [Vitis vinifera] |
| 14 |
Hb_004669_030 |
0.1548433796 |
- |
- |
- |
| 15 |
Hb_000215_200 |
0.1561581637 |
- |
- |
RecName: Full=Profilin-2; AltName: Full=Pollen allergen Hev b 8.0102; AltName: Allergen=Hev b 8.0102 [Hevea brasiliensis] |
| 16 |
Hb_000635_070 |
0.1584708347 |
- |
- |
PREDICTED: probable signal peptidase complex subunit 1 [Jatropha curcas] |
| 17 |
Hb_006153_030 |
0.160277966 |
- |
- |
PREDICTED: probable transaldolase [Jatropha curcas] |
| 18 |
Hb_003688_110 |
0.1607885897 |
- |
- |
- |
| 19 |
Hb_000125_020 |
0.1613086461 |
transcription factor |
TF Family: mTERF |
- |
| 20 |
Hb_080601_020 |
0.1616130813 |
- |
- |
PREDICTED: cytochrome c oxidase copper chaperone 1 [Jatropha curcas] |