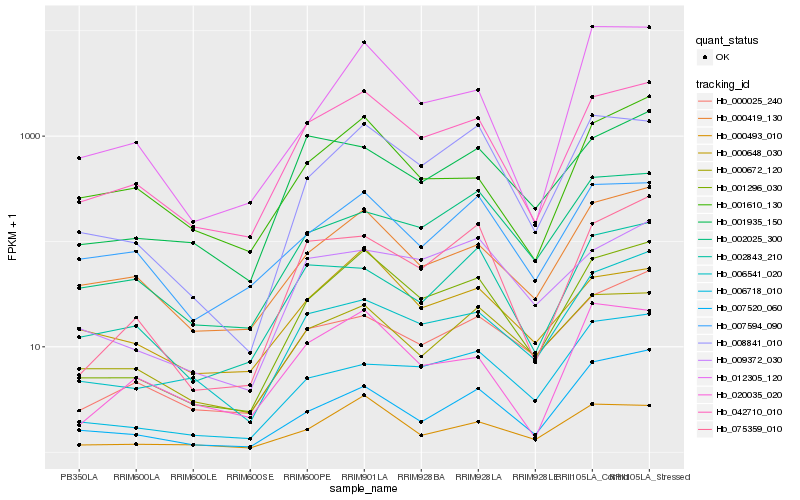
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001296_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638276 [Jatropha curcas] |
| 2 |
Hb_042710_010 |
0.0860404228 |
- |
- |
PREDICTED: 40S ribosomal protein S15a-1 [Jatropha curcas] |
| 3 |
Hb_008841_010 |
0.1142216428 |
- |
- |
PREDICTED: uncharacterized protein LOC103323653 [Prunus mume] |
| 4 |
Hb_000493_010 |
0.1293008974 |
- |
- |
hypothetical protein CICLE_v10021431mg [Citrus clementina] |
| 5 |
Hb_000419_130 |
0.1430363321 |
- |
- |
PREDICTED: uncharacterized protein LOC105650901 [Jatropha curcas] |
| 6 |
Hb_075359_010 |
0.1455822129 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 7 |
Hb_000025_240 |
0.1492134698 |
- |
- |
PREDICTED: uncharacterized protein LOC105628964 [Jatropha curcas] |
| 8 |
Hb_006541_020 |
0.1507919358 |
- |
- |
unknown [Lotus japonicus] |
| 9 |
Hb_007520_060 |
0.152379961 |
- |
- |
PREDICTED: putative exosome complex component rrp40 [Jatropha curcas] |
| 10 |
Hb_009372_030 |
0.1565872333 |
- |
- |
50S ribosomal protein L6, putative [Ricinus communis] |
| 11 |
Hb_012305_120 |
0.159749785 |
- |
- |
PREDICTED: 60S ribosomal protein L29-1-like [Glycine max] |
| 12 |
Hb_002025_300 |
0.1603207568 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 7 [Jatropha curcas] |
| 13 |
Hb_000672_120 |
0.165657162 |
- |
- |
V-type proton ATPase subunit G 1 [Jatropha curcas] |
| 14 |
Hb_006718_010 |
0.1677997218 |
- |
- |
PREDICTED: histone H2AX-like [Elaeis guineensis] |
| 15 |
Hb_000648_030 |
0.1683892606 |
- |
- |
hypothetical protein PHAVU_002G055100g [Phaseolus vulgaris] |
| 16 |
Hb_020035_020 |
0.1693601419 |
- |
- |
PREDICTED: uncharacterized protein LOC105631020 [Jatropha curcas] |
| 17 |
Hb_007594_090 |
0.170003169 |
- |
- |
PREDICTED: histone deacetylase complex subunit SAP18 [Jatropha curcas] |
| 18 |
Hb_001935_150 |
0.170743764 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit beta [Jatropha curcas] |
| 19 |
Hb_001610_130 |
0.1730351425 |
- |
- |
40S ribosomal protein S30 [Medicago truncatula] |
| 20 |
Hb_002843_210 |
0.175310702 |
- |
- |
PREDICTED: uncharacterized protein LOC105647178 [Jatropha curcas] |