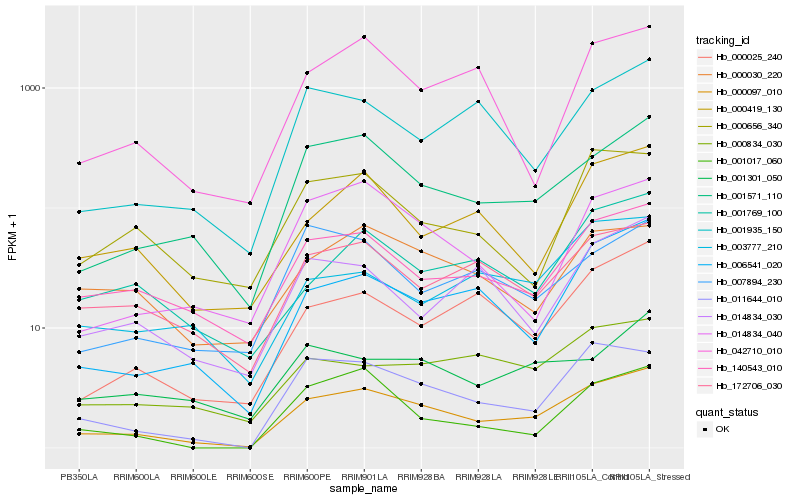
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000097_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_001571_110 |
0.1180272193 |
- |
- |
PREDICTED: 60S ribosomal protein L44-like [Gossypium raimondii] |
| 3 |
Hb_011644_010 |
0.1243530062 |
- |
- |
PREDICTED: probable choline-phosphate cytidylyltransferase [Elaeis guineensis] |
| 4 |
Hb_140543_010 |
0.1412851233 |
- |
- |
- |
| 5 |
Hb_006541_020 |
0.1468275487 |
- |
- |
unknown [Lotus japonicus] |
| 6 |
Hb_172706_030 |
0.1493847138 |
- |
- |
PREDICTED: uncharacterized protein LOC105640907 [Jatropha curcas] |
| 7 |
Hb_000025_240 |
0.1541044683 |
- |
- |
PREDICTED: uncharacterized protein LOC105628964 [Jatropha curcas] |
| 8 |
Hb_000419_130 |
0.1621174192 |
- |
- |
PREDICTED: uncharacterized protein LOC105650901 [Jatropha curcas] |
| 9 |
Hb_001935_150 |
0.1623533883 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit beta [Jatropha curcas] |
| 10 |
Hb_014834_040 |
0.1626940004 |
- |
- |
hypothetical protein JCGZ_03409 [Jatropha curcas] |
| 11 |
Hb_000834_030 |
0.1664027472 |
- |
- |
PREDICTED: protein Mpv17 isoform X2 [Jatropha curcas] |
| 12 |
Hb_001769_100 |
0.1679962618 |
- |
- |
hypothetical protein POPTR_0014s01620g [Populus trichocarpa] |
| 13 |
Hb_001017_060 |
0.1694876348 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 14 |
Hb_000030_220 |
0.1697141295 |
- |
- |
PREDICTED: uncharacterized protein LOC105650405 [Jatropha curcas] |
| 15 |
Hb_042710_010 |
0.1702424611 |
- |
- |
PREDICTED: 40S ribosomal protein S15a-1 [Jatropha curcas] |
| 16 |
Hb_001301_050 |
0.1744649125 |
- |
- |
- |
| 17 |
Hb_007894_230 |
0.1747498171 |
- |
- |
30S ribosomal protein S14, putative [Ricinus communis] |
| 18 |
Hb_000656_340 |
0.1751725431 |
- |
- |
hypothetical protein JCGZ_24686 [Jatropha curcas] |
| 19 |
Hb_003777_210 |
0.1765499628 |
- |
- |
thioredoxin m(mitochondrial)-type, putative [Ricinus communis] |
| 20 |
Hb_014834_030 |
0.1765542759 |
- |
- |
- |