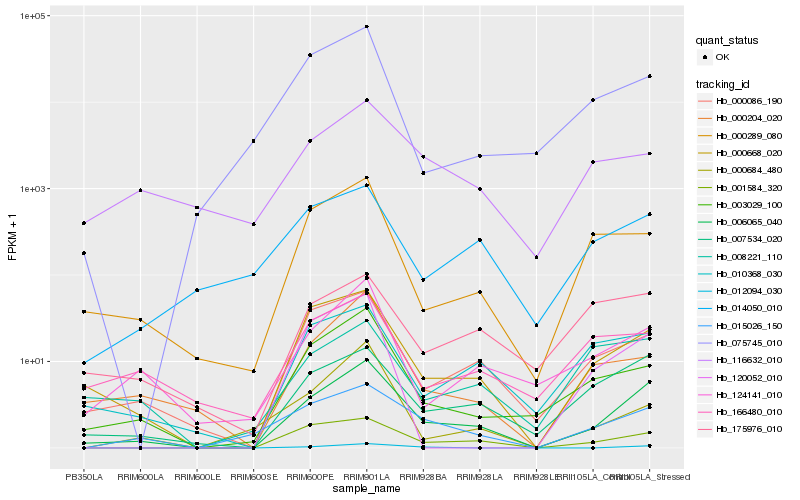
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006065_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_007534_020 |
0.1772518127 |
- |
- |
- |
| 3 |
Hb_000668_020 |
0.1776467556 |
- |
- |
- |
| 4 |
Hb_001584_320 |
0.1897795465 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 3 [Jatropha curcas] |
| 5 |
Hb_000086_190 |
0.1991993247 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 3-like protein [Cucumis sativus] |
| 6 |
Hb_003029_100 |
0.2093490122 |
- |
- |
- |
| 7 |
Hb_015026_150 |
0.2121476261 |
- |
- |
Nuclease PA3, putative [Ricinus communis] |
| 8 |
Hb_124141_010 |
0.2236794842 |
- |
- |
plant origin recognition complex subunit, putative [Ricinus communis] |
| 9 |
Hb_000204_020 |
0.2250166363 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized N-acetyltransferase ycf52 [Jatropha curcas] |
| 10 |
Hb_010368_030 |
0.2265982714 |
- |
- |
BnaA07g35570D [Brassica napus] |
| 11 |
Hb_000289_080 |
0.2267169583 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_012094_030 |
0.2281704899 |
- |
- |
PREDICTED: uncharacterized protein LOC103699727 [Phoenix dactylifera] |
| 13 |
Hb_014050_010 |
0.2487518184 |
- |
- |
PREDICTED: histone H3.3-like, partial [Phoenix dactylifera] |
| 14 |
Hb_175976_010 |
0.2503755938 |
- |
- |
- |
| 15 |
Hb_008221_110 |
0.2507723612 |
- |
- |
PREDICTED: diphthamide biosynthesis protein 4 [Jatropha curcas] |
| 16 |
Hb_000684_480 |
0.2564352948 |
transcription factor |
TF Family: Orphans |
- |
| 17 |
Hb_116632_010 |
0.2567169162 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_075745_010 |
0.2574220284 |
- |
- |
hypothetical protein EUGRSUZ_D00579 [Eucalyptus grandis] |
| 19 |
Hb_166480_010 |
0.263908447 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase, mitochondrial-like [Cucumis melo] |
| 20 |
Hb_120052_010 |
0.2642739668 |
- |
- |
hypothetical protein PRUPE_ppa1027171mg [Prunus persica] |