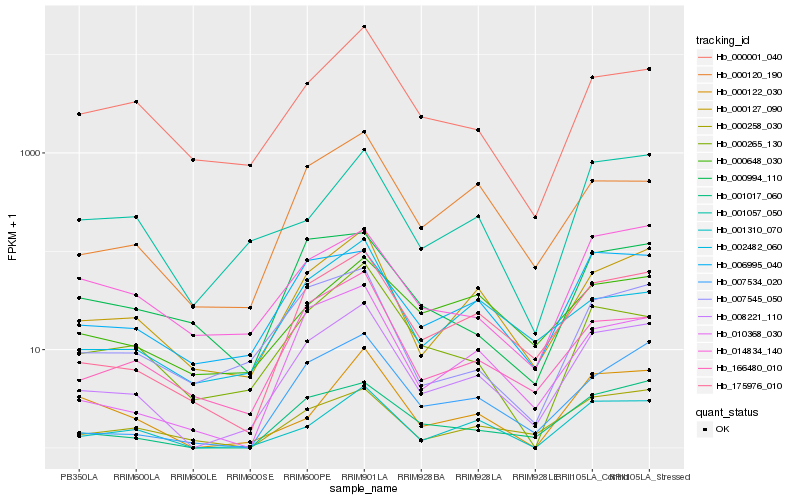
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008221_110 |
0.0 |
- |
- |
PREDICTED: diphthamide biosynthesis protein 4 [Jatropha curcas] |
| 2 |
Hb_175976_010 |
0.1083281353 |
- |
- |
- |
| 3 |
Hb_000127_090 |
0.1146449918 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001017_060 |
0.1351947234 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 5 |
Hb_001310_070 |
0.1470341093 |
- |
- |
PREDICTED: uncharacterized protein LOC105639838 [Jatropha curcas] |
| 6 |
Hb_010368_030 |
0.1503956038 |
- |
- |
BnaA07g35570D [Brassica napus] |
| 7 |
Hb_007534_020 |
0.1522874096 |
- |
- |
- |
| 8 |
Hb_006995_040 |
0.1529678505 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: 14 kDa zinc-binding protein-like [Pyrus x bretschneideri] |
| 9 |
Hb_166480_010 |
0.154878306 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase, mitochondrial-like [Cucumis melo] |
| 10 |
Hb_007545_050 |
0.1606863424 |
- |
- |
- |
| 11 |
Hb_000120_190 |
0.1624109317 |
- |
- |
Pre-mRNA-splicing factor ini1 [Ricinus communis] |
| 12 |
Hb_000258_030 |
0.1636859773 |
- |
- |
- |
| 13 |
Hb_000001_040 |
0.1685002697 |
- |
- |
unnamed protein product [Coffea canephora] |
| 14 |
Hb_000265_130 |
0.1759260001 |
- |
- |
- |
| 15 |
Hb_014834_140 |
0.1776404882 |
- |
- |
Uncharacterized protein TCM_015942 [Theobroma cacao] |
| 16 |
Hb_001057_050 |
0.1820087171 |
- |
- |
hypothetical protein POPTR_0003s15960g, partial [Populus trichocarpa] |
| 17 |
Hb_000122_030 |
0.1842097508 |
- |
- |
PREDICTED: uncharacterized protein LOC104606081, partial [Nelumbo nucifera] |
| 18 |
Hb_000994_110 |
0.1868030927 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002482_060 |
0.1880882224 |
- |
- |
hypothetical protein CICLE_v10033884mg, partial [Citrus clementina] |
| 20 |
Hb_000648_030 |
0.1897784366 |
- |
- |
hypothetical protein PHAVU_002G055100g [Phaseolus vulgaris] |