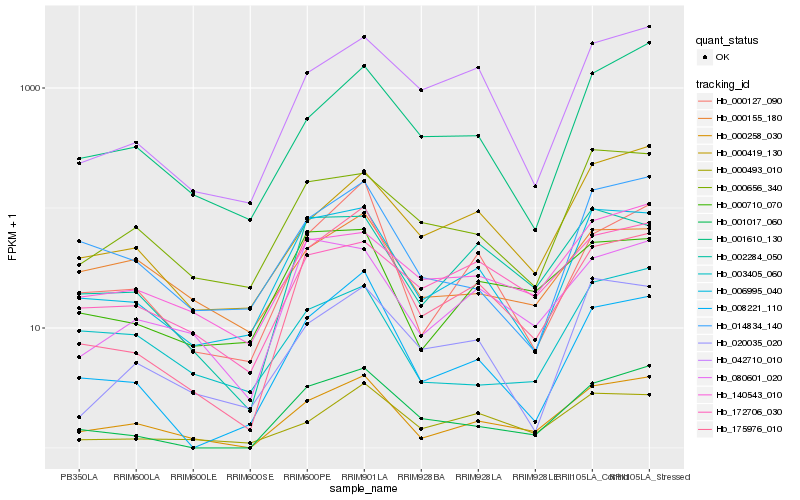
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000258_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_175976_010 |
0.1453713084 |
- |
- |
- |
| 3 |
Hb_000127_090 |
0.1475245288 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_006995_040 |
0.1481935425 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: 14 kDa zinc-binding protein-like [Pyrus x bretschneideri] |
| 5 |
Hb_001017_060 |
0.1597448058 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 6 |
Hb_002284_050 |
0.1620083656 |
- |
- |
hypothetical protein POPTR_0002s00740g [Populus trichocarpa] |
| 7 |
Hb_008221_110 |
0.1636859773 |
- |
- |
PREDICTED: diphthamide biosynthesis protein 4 [Jatropha curcas] |
| 8 |
Hb_080601_020 |
0.1637300758 |
- |
- |
PREDICTED: cytochrome c oxidase copper chaperone 1 [Jatropha curcas] |
| 9 |
Hb_000419_130 |
0.1640306496 |
- |
- |
PREDICTED: uncharacterized protein LOC105650901 [Jatropha curcas] |
| 10 |
Hb_001610_130 |
0.1645457604 |
- |
- |
40S ribosomal protein S30 [Medicago truncatula] |
| 11 |
Hb_014834_140 |
0.1653903432 |
- |
- |
Uncharacterized protein TCM_015942 [Theobroma cacao] |
| 12 |
Hb_003405_060 |
0.165936257 |
- |
- |
- |
| 13 |
Hb_140543_010 |
0.1700657163 |
- |
- |
- |
| 14 |
Hb_000493_010 |
0.1714063585 |
- |
- |
hypothetical protein CICLE_v10021431mg [Citrus clementina] |
| 15 |
Hb_000710_070 |
0.1732284161 |
- |
- |
PREDICTED: uncharacterized protein LOC105645553 [Jatropha curcas] |
| 16 |
Hb_000656_340 |
0.1751468627 |
- |
- |
hypothetical protein JCGZ_24686 [Jatropha curcas] |
| 17 |
Hb_172706_030 |
0.178649317 |
- |
- |
PREDICTED: uncharacterized protein LOC105640907 [Jatropha curcas] |
| 18 |
Hb_020035_020 |
0.1850148921 |
- |
- |
PREDICTED: uncharacterized protein LOC105631020 [Jatropha curcas] |
| 19 |
Hb_042710_010 |
0.1866841376 |
- |
- |
PREDICTED: 40S ribosomal protein S15a-1 [Jatropha curcas] |
| 20 |
Hb_000155_180 |
0.1886488914 |
- |
- |
PREDICTED: 28 kDa heat- and acid-stable phosphoprotein [Jatropha curcas] |