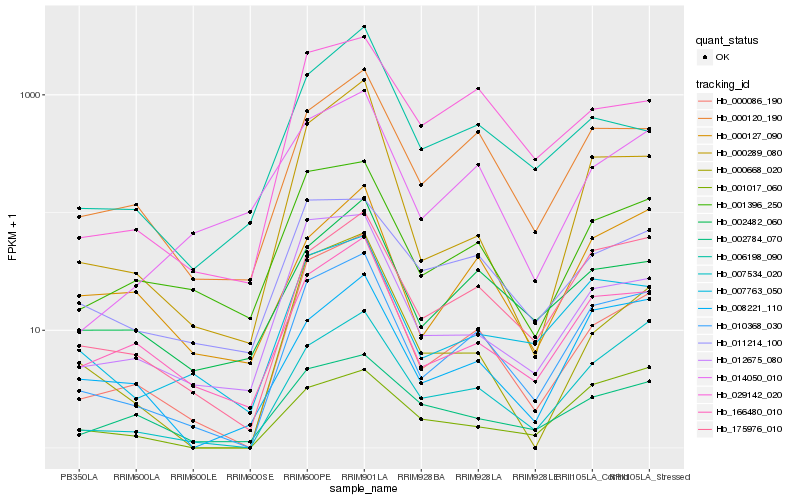
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010368_030 |
0.0 |
- |
- |
BnaA07g35570D [Brassica napus] |
| 2 |
Hb_175976_010 |
0.0998767176 |
- |
- |
- |
| 3 |
Hb_000086_190 |
0.117370658 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 3-like protein [Cucumis sativus] |
| 4 |
Hb_000120_190 |
0.1225393884 |
- |
- |
Pre-mRNA-splicing factor ini1 [Ricinus communis] |
| 5 |
Hb_007534_020 |
0.1256835347 |
- |
- |
- |
| 6 |
Hb_007763_050 |
0.1343189716 |
- |
- |
hypothetical protein L484_012752 [Morus notabilis] |
| 7 |
Hb_166480_010 |
0.1420114888 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase, mitochondrial-like [Cucumis melo] |
| 8 |
Hb_001396_250 |
0.1461237441 |
- |
- |
PREDICTED: stress-associated endoplasmic reticulum protein 2 [Jatropha curcas] |
| 9 |
Hb_008221_110 |
0.1503956038 |
- |
- |
PREDICTED: diphthamide biosynthesis protein 4 [Jatropha curcas] |
| 10 |
Hb_002482_060 |
0.1602651251 |
- |
- |
hypothetical protein CICLE_v10033884mg, partial [Citrus clementina] |
| 11 |
Hb_000127_090 |
0.1643736988 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000289_080 |
0.1647340113 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_029142_020 |
0.1671488581 |
- |
- |
hypothetical protein POPTR_0004s23900g [Populus trichocarpa] |
| 14 |
Hb_011214_100 |
0.1696117699 |
- |
- |
PREDICTED: uncharacterized protein LOC105633003 [Jatropha curcas] |
| 15 |
Hb_014050_010 |
0.1724821024 |
- |
- |
PREDICTED: histone H3.3-like, partial [Phoenix dactylifera] |
| 16 |
Hb_000668_020 |
0.1745848591 |
- |
- |
- |
| 17 |
Hb_012675_080 |
0.1749714813 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 1 [Populus euphratica] |
| 18 |
Hb_002784_070 |
0.1837447051 |
- |
- |
BnaUnng01470D [Brassica napus] |
| 19 |
Hb_001017_060 |
0.1850235324 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 20 |
Hb_006198_090 |
0.1949361554 |
- |
- |
PREDICTED: vesicle-associated membrane protein 721-like isoform X2 [Tarenaya hassleriana] |