| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
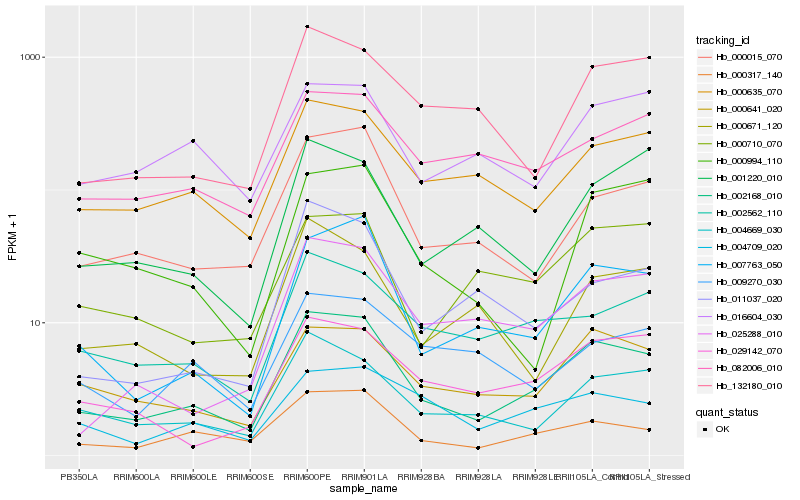
Hb_002168_010 |
0.0 |
- |
- |
PREDICTED: bifunctional dihydrofolate reductase-thymidylate synthase-like isoform X4 [Vitis vinifera] |
| 2 |
Hb_000317_140 |
0.1268020175 |
- |
- |
hypothetical protein CISIN_1g0019781mg, partial [Citrus sinensis] |
| 3 |
Hb_007763_050 |
0.1336335395 |
- |
- |
hypothetical protein L484_012752 [Morus notabilis] |
| 4 |
Hb_004669_030 |
0.1347733731 |
- |
- |
- |
| 5 |
Hb_000641_020 |
0.1377895791 |
- |
- |
PREDICTED: tubulin-folding cofactor B-like [Jatropha curcas] |
| 6 |
Hb_000015_070 |
0.1430218584 |
- |
- |
hypothetical protein POPTR_0001s22690g [Populus trichocarpa] |
| 7 |
Hb_000635_070 |
0.1450173184 |
- |
- |
PREDICTED: probable signal peptidase complex subunit 1 [Jatropha curcas] |
| 8 |
Hb_002562_110 |
0.1511602698 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX3, mitochondrial [Jatropha curcas] |
| 9 |
Hb_029142_070 |
0.1530703823 |
- |
- |
- |
| 10 |
Hb_132180_010 |
0.1606883233 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 11 |
Hb_082006_010 |
0.1615274621 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004709_020 |
0.1616244483 |
- |
- |
PREDICTED: putative threonine aspartase [Jatropha curcas] |
| 13 |
Hb_025288_010 |
0.1652462027 |
- |
- |
PREDICTED: protein unc-50 homolog [Jatropha curcas] |
| 14 |
Hb_000710_070 |
0.1680154244 |
- |
- |
PREDICTED: uncharacterized protein LOC105645553 [Jatropha curcas] |
| 15 |
Hb_016604_030 |
0.1712160075 |
- |
- |
histone H4 [Zea mays] |
| 16 |
Hb_001220_010 |
0.1734540222 |
- |
- |
hypothetical protein JCGZ_00411 [Jatropha curcas] |
| 17 |
Hb_000671_120 |
0.1734895652 |
- |
- |
hypothetical protein JCGZ_23777 [Jatropha curcas] |
| 18 |
Hb_000994_110 |
0.1759707963 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_011037_020 |
0.1779697995 |
- |
- |
unknown [Picea sitchensis] |
| 20 |
Hb_009270_030 |
0.1802309993 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |