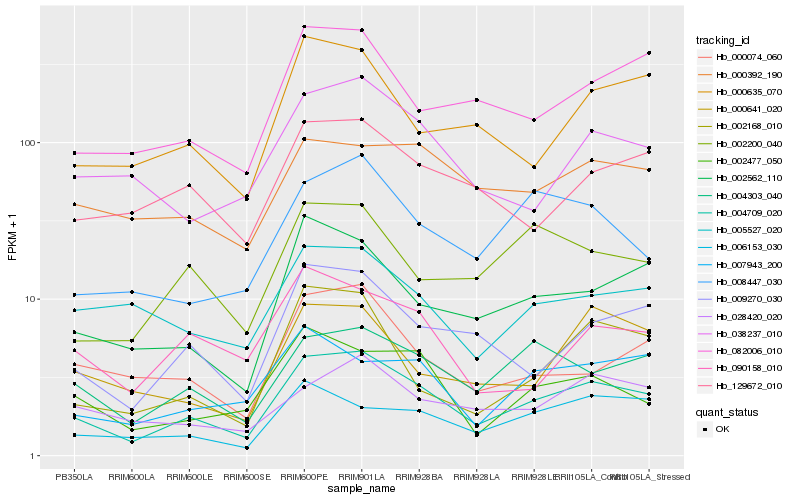
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004709_020 |
0.0 |
- |
- |
PREDICTED: putative threonine aspartase [Jatropha curcas] |
| 2 |
Hb_004303_040 |
0.1356519848 |
- |
- |
PREDICTED: fumarate hydratase 1, mitochondrial [Jatropha curcas] |
| 3 |
Hb_009270_030 |
0.1380755861 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 4 |
Hb_002562_110 |
0.1404132629 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX3, mitochondrial [Jatropha curcas] |
| 5 |
Hb_082006_010 |
0.1502483618 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_008447_030 |
0.1524942027 |
- |
- |
- |
| 7 |
Hb_038237_010 |
0.1531184502 |
- |
- |
hypothetical protein POPTR_0003s01110g [Populus trichocarpa] |
| 8 |
Hb_002477_050 |
0.1533067354 |
- |
- |
unnamed protein product [Coffea canephora] |
| 9 |
Hb_000074_060 |
0.1573353451 |
- |
- |
PREDICTED: membrane-associated progesterone-binding protein 4-like [Populus euphratica] |
| 10 |
Hb_002200_040 |
0.1575367587 |
- |
- |
PREDICTED: uncharacterized protein LOC105629143 isoform X1 [Jatropha curcas] |
| 11 |
Hb_129672_010 |
0.1579963089 |
- |
- |
PREDICTED: signal peptidase complex subunit 3B [Jatropha curcas] |
| 12 |
Hb_006153_030 |
0.1585062228 |
- |
- |
PREDICTED: probable transaldolase [Jatropha curcas] |
| 13 |
Hb_028420_020 |
0.1588157676 |
- |
- |
PREDICTED: uncharacterized protein LOC105647566 [Jatropha curcas] |
| 14 |
Hb_000392_190 |
0.1591359882 |
- |
- |
PREDICTED: NAP1-related protein 2 [Jatropha curcas] |
| 15 |
Hb_000635_070 |
0.1600299929 |
- |
- |
PREDICTED: probable signal peptidase complex subunit 1 [Jatropha curcas] |
| 16 |
Hb_007943_200 |
0.1605715403 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 17 |
Hb_005527_020 |
0.1613785553 |
- |
- |
PREDICTED: uncharacterized protein LOC105643027 [Jatropha curcas] |
| 18 |
Hb_002168_010 |
0.1616244483 |
- |
- |
PREDICTED: bifunctional dihydrofolate reductase-thymidylate synthase-like isoform X4 [Vitis vinifera] |
| 19 |
Hb_000641_020 |
0.1627430224 |
- |
- |
PREDICTED: tubulin-folding cofactor B-like [Jatropha curcas] |
| 20 |
Hb_090158_010 |
0.163586467 |
- |
- |
hypothetical protein RCOM_1968400 [Ricinus communis] |