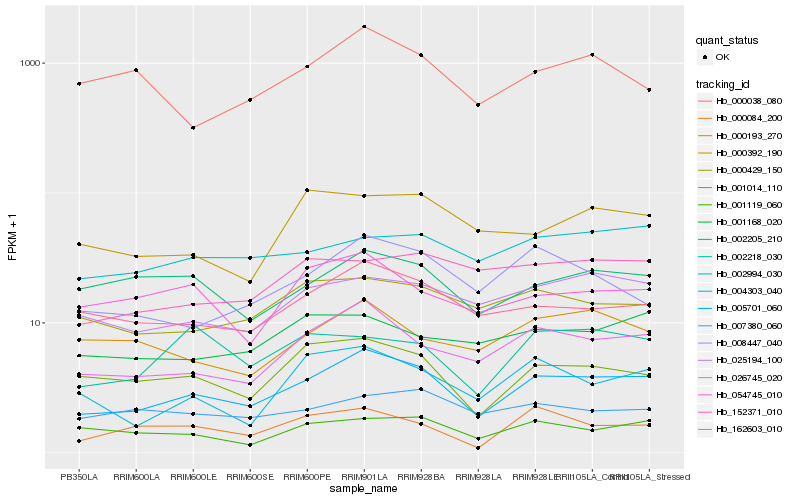
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005701_060 |
0.0 |
- |
- |
hypothetical protein JCGZ_04513 [Jatropha curcas] |
| 2 |
Hb_026745_020 |
0.1019993345 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 8, chloroplastic [Jatropha curcas] |
| 3 |
Hb_162603_010 |
0.1218287284 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 4 |
Hb_001014_110 |
0.1239644216 |
- |
- |
PREDICTED: ubiquitin thioesterase otubain-like [Jatropha curcas] |
| 5 |
Hb_025194_100 |
0.1314202841 |
- |
- |
PREDICTED: probable 2-oxoglutarate/Fe(II)-dependent dioxygenase [Jatropha curcas] |
| 6 |
Hb_000429_150 |
0.1400340087 |
- |
- |
hypothetical protein JCGZ_06799 [Jatropha curcas] |
| 7 |
Hb_004303_040 |
0.1414727643 |
- |
- |
PREDICTED: fumarate hydratase 1, mitochondrial [Jatropha curcas] |
| 8 |
Hb_008447_040 |
0.142222377 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 9 |
Hb_002205_210 |
0.1422231757 |
- |
- |
PREDICTED: aminoacyl tRNA synthase complex-interacting multifunctional protein 1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_002994_030 |
0.145627658 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM22-like [Pyrus x bretschneideri] |
| 11 |
Hb_054745_010 |
0.1466152344 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001119_060 |
0.1473587289 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X3 [Malus domestica] |
| 13 |
Hb_000084_200 |
0.1493135714 |
- |
- |
PREDICTED: uncharacterized protein LOC105634688 isoform X2 [Jatropha curcas] |
| 14 |
Hb_152371_010 |
0.1493289648 |
- |
- |
RNA-binding KH domain-containing protein [Theobroma cacao] |
| 15 |
Hb_001168_020 |
0.150106068 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit gamma [Jatropha curcas] |
| 16 |
Hb_000392_190 |
0.15050188 |
- |
- |
PREDICTED: NAP1-related protein 2 [Jatropha curcas] |
| 17 |
Hb_000038_080 |
0.1509884584 |
- |
- |
PREDICTED: glutathione S-transferase F9-like [Jatropha curcas] |
| 18 |
Hb_000193_270 |
0.1514776292 |
transcription factor |
TF Family: Pseudo ARR-B |
PREDICTED: two-component response regulator-like APRR1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_007380_060 |
0.1518982572 |
- |
- |
hypothetical protein POPTR_0006s00280g [Populus trichocarpa] |
| 20 |
Hb_002218_030 |
0.152409022 |
- |
- |
hypothetical protein B456_007G345200 [Gossypium raimondii] |