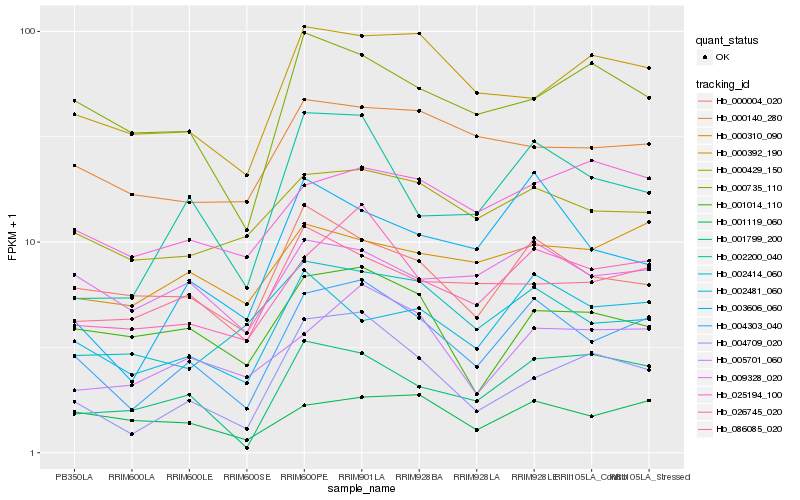
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004303_040 |
0.0 |
- |
- |
PREDICTED: fumarate hydratase 1, mitochondrial [Jatropha curcas] |
| 2 |
Hb_026745_020 |
0.1098974507 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 8, chloroplastic [Jatropha curcas] |
| 3 |
Hb_002481_060 |
0.1354434117 |
- |
- |
PREDICTED: uncharacterized protein LOC105640851 [Jatropha curcas] |
| 4 |
Hb_004709_020 |
0.1356519848 |
- |
- |
PREDICTED: putative threonine aspartase [Jatropha curcas] |
| 5 |
Hb_002200_040 |
0.1382224322 |
- |
- |
PREDICTED: uncharacterized protein LOC105629143 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000004_020 |
0.140787597 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_005701_060 |
0.1414727643 |
- |
- |
hypothetical protein JCGZ_04513 [Jatropha curcas] |
| 8 |
Hb_001119_060 |
0.142954601 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X3 [Malus domestica] |
| 9 |
Hb_003606_060 |
0.1434525567 |
- |
- |
PREDICTED: uncharacterized protein LOC105649867 [Jatropha curcas] |
| 10 |
Hb_000429_150 |
0.1457121905 |
- |
- |
hypothetical protein JCGZ_06799 [Jatropha curcas] |
| 11 |
Hb_001799_200 |
0.1466834821 |
- |
- |
PREDICTED: tRNA wybutosine-synthesizing protein 4 [Jatropha curcas] |
| 12 |
Hb_000392_190 |
0.1512643955 |
- |
- |
PREDICTED: NAP1-related protein 2 [Jatropha curcas] |
| 13 |
Hb_009328_020 |
0.1513296136 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002414_060 |
0.1550997042 |
- |
- |
hypothetical protein EUGRSUZ_E01310 [Eucalyptus grandis] |
| 15 |
Hb_000735_110 |
0.1552840203 |
- |
- |
fk506-binding protein, putative [Ricinus communis] |
| 16 |
Hb_001014_110 |
0.1561513719 |
- |
- |
PREDICTED: ubiquitin thioesterase otubain-like [Jatropha curcas] |
| 17 |
Hb_086085_020 |
0.1564187475 |
- |
- |
Non-imprinted in Prader-Willi/Angelman syndrome region protein isoform 3 [Theobroma cacao] |
| 18 |
Hb_025194_100 |
0.1564661651 |
- |
- |
PREDICTED: probable 2-oxoglutarate/Fe(II)-dependent dioxygenase [Jatropha curcas] |
| 19 |
Hb_000140_280 |
0.1568232048 |
- |
- |
hypothetical protein B456_001G157900 [Gossypium raimondii] |
| 20 |
Hb_000310_090 |
0.1582337326 |
- |
- |
3-oxoacyl-[acyl-carrier-protein] synthase 3 A, chloroplastic [Jatropha curcas] |