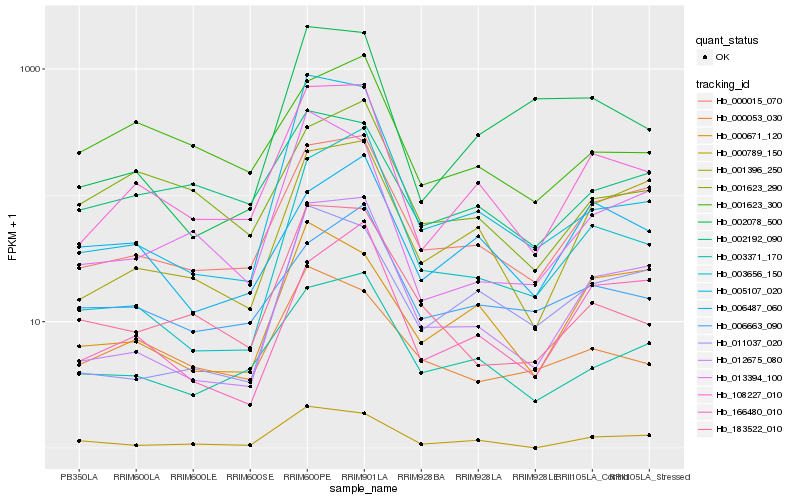
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_108227_010 |
0.0 |
- |
- |
RecName: Full=Profilin-2; AltName: Full=Pollen allergen Hev b 8.0102; AltName: Allergen=Hev b 8.0102 [Hevea brasiliensis] |
| 2 |
Hb_000015_070 |
0.1207890788 |
- |
- |
hypothetical protein POPTR_0001s22690g [Populus trichocarpa] |
| 3 |
Hb_003371_170 |
0.1409836252 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001396_250 |
0.1477460147 |
- |
- |
PREDICTED: stress-associated endoplasmic reticulum protein 2 [Jatropha curcas] |
| 5 |
Hb_012675_080 |
0.1490546552 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 1 [Populus euphratica] |
| 6 |
Hb_000789_150 |
0.1620860647 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_013394_100 |
0.1683931188 |
- |
- |
PREDICTED: uncharacterized protein LOC105135144 [Populus euphratica] |
| 8 |
Hb_002192_090 |
0.1701531909 |
- |
- |
hypothetical protein AMTR_s00029p00055000 [Amborella trichopoda] |
| 9 |
Hb_001623_290 |
0.1744750037 |
- |
- |
unknown [Populus trichocarpa] |
| 10 |
Hb_183522_010 |
0.1759624049 |
- |
- |
Far upstream element-binding protein, putative [Ricinus communis] |
| 11 |
Hb_166480_010 |
0.1773754336 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase, mitochondrial-like [Cucumis melo] |
| 12 |
Hb_005107_020 |
0.1791782344 |
- |
- |
hypothetical protein JCGZ_25266 [Jatropha curcas] |
| 13 |
Hb_011037_020 |
0.1801566053 |
- |
- |
unknown [Picea sitchensis] |
| 14 |
Hb_001623_300 |
0.1806481715 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 13-B [Jatropha curcas] |
| 15 |
Hb_000671_120 |
0.1809421324 |
- |
- |
hypothetical protein JCGZ_23777 [Jatropha curcas] |
| 16 |
Hb_006663_090 |
0.1834552745 |
- |
- |
hypothetical protein B456_010G1255001, partial [Gossypium raimondii] |
| 17 |
Hb_003656_150 |
0.1846145709 |
- |
- |
- |
| 18 |
Hb_006487_060 |
0.1863206199 |
- |
- |
Defective in cullin neddylation protein, putative [Ricinus communis] |
| 19 |
Hb_000053_030 |
0.1864417583 |
- |
- |
hypothetical protein CISIN_1g0010393mg, partial [Citrus sinensis] |
| 20 |
Hb_002078_500 |
0.1865898676 |
- |
- |
conserved hypothetical protein [Ricinus communis] |